Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination
- PMID: 10601032
- PMCID: PMC317185
- DOI: 10.1101/gad.13.23.3059
Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination
Abstract
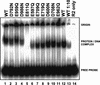
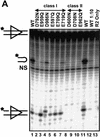
RAG1 and RAG2 initiate V(D)J recombination, the process of rearranging the antigen-binding domain of immunoglobulins and T-cell receptors, by introducing site-specific double-strand breaks (DSB) in chromosomal DNA during lymphocyte development. These breaks are generated in two steps, nicking of one strand (hydrolysis), followed by hairpin formation (transesterification). The nature and location of the RAG active site(s) have remained unknown. Because acidic amino acids have a critical role in catalyzing DNA cleavage by nucleases and recombinases that require divalent metal ions as cofactors, we hypothesized that acidic active site residues are likewise essential for RAG-mediated DNA cleavage. We altered each conserved acidic amino acid in RAG1 and RAG2 by site-directed mutagenesis, and examined >100 mutants using a combination of in vivo and in vitro analyses. No conserved acidic amino acids in RAG2 were critical for catalysis; three RAG1 mutants retained normal DNA binding, but were catalytically inactive for both nicking and hairpin formation. These data argue that one active site in RAG1 performs both steps of the cleavage reaction. Amino acid substitution experiments that changed the metal ion specificity suggest that at least one of these three residues contacts the metal ion(s) directly. These data suggest that RAG-mediated DNA cleavage involves coordination of divalent metal ion(s) by RAG1.
Figures

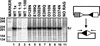






Similar articles
-
Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase.Genes Dev. 1999 Dec 1;13(23):3070-80. doi: 10.1101/gad.13.23.3070. Genes Dev. 1999. PMID: 10601033 Free PMC article.
-
Understanding how the V(D)J recombinase catalyzes transesterification: distinctions between DNA cleavage and transposition.Nucleic Acids Res. 2008 May;36(9):2864-73. doi: 10.1093/nar/gkn128. Epub 2008 Mar 29. Nucleic Acids Res. 2008. PMID: 18375979 Free PMC article.
-
The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination.Mol Cell Biol. 2001 Jan;21(2):449-58. doi: 10.1128/MCB.21.2.449-458.2001. Mol Cell Biol. 2001. PMID: 11134333 Free PMC article.
-
The RAG proteins and V(D)J recombination: complexes, ends, and transposition.Annu Rev Immunol. 2000;18:495-527. doi: 10.1146/annurev.immunol.18.1.495. Annu Rev Immunol. 2000. PMID: 10837067 Review.
-
RAG1 and RAG2 in V(D)J recombination and transposition.Immunol Res. 2001;23(1):23-39. doi: 10.1385/IR:23:1:23. Immunol Res. 2001. PMID: 11417858 Review.
Cited by
-
P Transposable Elements in Drosophila and other Eukaryotic Organisms.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0004-2014. doi: 10.1128/microbiolspec.MDNA3-0004-2014. Microbiol Spectr. 2015. PMID: 26104714 Free PMC article. Review.
-
Retroviral Integrase Structure and DNA Recombination Mechanism.Microbiol Spectr. 2014;2(6):1-22. doi: 10.1128/microbiolspec.MDNA3-0024-2014.. Microbiol Spectr. 2014. PMID: 25705574 Free PMC article.
-
Footprint analysis of recombination signal sequences in the 12/23 synaptic complex of V(D)J recombination.Mol Cell Biol. 2002 Oct;22(20):7217-25. doi: 10.1128/MCB.22.20.7217-7225.2002. Mol Cell Biol. 2002. PMID: 12242298 Free PMC article.
-
Engineered Sleeping Beauty transposase redirects transposon integration away from genes.Nucleic Acids Res. 2022 Mar 21;50(5):2807-2825. doi: 10.1093/nar/gkac092. Nucleic Acids Res. 2022. PMID: 35188569 Free PMC article.
-
Overlapping signals for protein degradation and nuclear localization define a role for intrinsic RAG-2 nuclear uptake in dividing cells.Mol Cell Biol. 2003 Aug;23(15):5308-19. doi: 10.1128/MCB.23.15.5308-5319.2003. Mol Cell Biol. 2003. PMID: 12861017 Free PMC article.
References
-
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. - PubMed
-
- Allingham JS, Pribil PA, Haniford DB. All three residues of the Tn10 transposase DDE catalytic triad function in divalent metal ion binding. J Mol Biol. 1999;289:1195–1206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
