Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles
- PMID: 10564255
- PMCID: PMC25634
- DOI: 10.1091/mbc.10.11.3549
Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles
Abstract
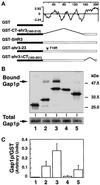
The SHR3 gene of Saccharomyces cerevisiae encodes an integral membrane component of the endoplasmic reticulum (ER) with four membrane-spanning segments and a hydrophilic, cytoplasmically oriented carboxyl-terminal domain. Mutations in SHR3 specifically impede the transport of all 18 members of the amino acid permease (aap) gene family away from the ER. Shr3p does not itself exit the ER. Aaps fully integrate into the ER membrane and fold properly independently of Shr3p. Shr3p physically associates with the general aap Gap1p but not Sec61p, Gal2p, or Pma1p in a complex that can be purified from N-dodecylmaltoside-solubilized membranes. Pulse-chase experiments indicate that the Shr3p-Gap1p association is transient, a reflection of the exit of Gap1p from the ER. The ER-derived vesicle COPII coatomer components Sec13p, Sec23p, Sec24p, and Sec31p but not Sar1p bind Shr3p via interactions with its carboxyl-terminal domain. The mutant shr3-23p, a nonfunctional membrane-associated protein, is unable to associate with aaps but retains the capacity to bind COPII components. The overexpression of either Shr3p or shr3-23p partially suppresses the temperature-sensitive sec12-1 allele. These results are consistent with a model in which Shr3p acts as a packaging chaperone that initiates ER-derived transport vesicle formation in the proximity of aaps by facilitating the membrane association and assembly of COPII coatomer components.
Figures





Similar articles
-
Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro.J Cell Biol. 1996 Nov;135(3):585-95. doi: 10.1083/jcb.135.3.585. J Cell Biol. 1996. PMID: 8909535 Free PMC article.
-
Selective packaging of cargo molecules into endoplasmic reticulum-derived COPII vesicles.Proc Natl Acad Sci U S A. 1997 Feb 4;94(3):837-42. doi: 10.1073/pnas.94.3.837. Proc Natl Acad Sci U S A. 1997. PMID: 9023343 Free PMC article.
-
COPII-cargo interactions direct protein sorting into ER-derived transport vesicles.Nature. 1998 Jan 8;391(6663):187-90. doi: 10.1038/34438. Nature. 1998. PMID: 9428766
-
SHR3 function is linked to cOPII mediated ER vesicle formation.Folia Microbiol (Praha). 1996;41(1):93. doi: 10.1007/BF02816353. Folia Microbiol (Praha). 1996. PMID: 9090835 No abstract available.
-
COPII: a membrane coat that forms endoplasmic reticulum-derived vesicles.FEBS Lett. 1995 Aug 1;369(1):93-6. doi: 10.1016/0014-5793(95)00618-j. FEBS Lett. 1995. PMID: 7641893 Review.
Cited by
-
Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae.Genetics. 2004 Dec;168(4):1899-913. doi: 10.1534/genetics.104.032961. Epub 2004 Sep 15. Genetics. 2004. PMID: 15371354 Free PMC article.
-
Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3.J Cell Biol. 2006 Jul 3;174(1):19-25. doi: 10.1083/jcb.200602049. J Cell Biol. 2006. PMID: 16818716 Free PMC article.
-
Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer.J Cell Biol. 2001 Mar 5;152(5):935-44. doi: 10.1083/jcb.152.5.935. J Cell Biol. 2001. PMID: 11238450 Free PMC article.
-
The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast.Chem Phys Lipids. 2014 May;180:23-43. doi: 10.1016/j.chemphyslip.2013.12.013. Epub 2014 Jan 10. Chem Phys Lipids. 2014. PMID: 24418527 Free PMC article. Review.
-
Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids.Mol Cell Biol. 2001 Feb;21(3):814-26. doi: 10.1128/MCB.21.3.814-826.2001. Mol Cell Biol. 2001. PMID: 11154269 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

