The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10)
- PMID: 10559364
- PMCID: PMC113101
- DOI: 10.1128/JVI.73.12.10458-10471.1999
The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10)
Abstract
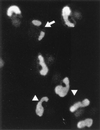
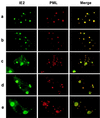
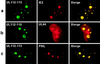
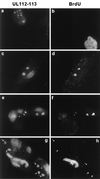
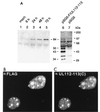
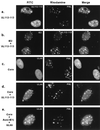
During human cytomegalovirus (HCMV) infection, the periphery of promyelocytic leukemia protein (PML)-associated nuclear bodies (also known as PML oncogenic domains [PODs] or ND10) are sites for both input viral genome deposition and immediate-early (IE) gene transcription. At very early times after infection, the IE1 protein localizes to and subsequently disrupts PODs, whereas the IE2 protein localizes within or adjacent to PODs. This process appears to be required for efficient viral gene expression and DNA replication. We have investigated the initiation of viral DNA replication compartment formation by studying the localization of viral IE proteins, DNA replication proteins, and the PML protein during productive infection. Localization of IE2 adjacent to PODs between 2 and 6 h after infection was confirmed by confocal microscopy of human fibroblasts (HF cells) infected with both wild-type HCMV(Towne) and with an IE1-deletion mutant HCMV(CR208) that fails to disrupt PODs. In HCMV(Towne)-infected HF cells at 24 to 48 h, IE2 also accumulated in newly formed viral DNA replication compartments containing the polymerase processivity factor (UL44), the single-stranded DNA binding protein (SSB; UL57), the UL112-113 accessory protein, and newly incorporated bromodeoxyuridine (BrdU). Double labeling of the HCMV(CR208)-infected HF cells demonstrated that formation of viral DNA replication compartments initiates within granular structures that bud from the periphery of some of the PODs and subsequently coalesce into larger structures that are flanked by PODs. In transient DNA transfection assays, both the N terminus (codons 136 to 290) and the C terminus (codons 379 to 579) of IE2 exon 5, but not the central region between them, were found to be necessary for both the punctate distribution of IE2 and its association with PODs. Like IE2, the UL112-113 accessory replication protein was also distributed in a POD-associated pattern in both DNA-transfected and virus-infected cells beginning at 6 h. Furthermore, when all six replication core machinery proteins (polymerase complex, SSB, and helicase-primase complex) were expressed together in the presence of UL112-113, they also accumulated at POD-associated sites, suggesting that the UL112-113 protein (but not IE2) may play a role in recruitment of viral replication fork proteins into the periphery of PODs. These results show that (i) subsequent to accumulating at the periphery of PODs, IE2 is incorporated together with the core proteins into viral DNA replication compartments that initiate from the periphery of PODs and then grow to fill the space between groups of PODs, and (ii) the UL112-113 protein appears to have a key role in assembling and recruiting the core replication machinery proteins in the initial stages of viral replication compartment formation.
Figures




Similar articles
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells.J Virol. 1997 Jun;71(6):4599-613. doi: 10.1128/JVI.71.6.4599-4613.1997. J Virol. 1997. PMID: 9151854 Free PMC article.
-
Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells.J Virol. 2007 Sep;81(18):10123-36. doi: 10.1128/JVI.01009-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626080 Free PMC article.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
-
Intrinsic Immune Mechanisms Restricting Human Cytomegalovirus Replication.Viruses. 2021 Jan 26;13(2):179. doi: 10.3390/v13020179. Viruses. 2021. PMID: 33530304 Free PMC article. Review.
Cited by
-
Requirement of the N-terminal residues of human cytomegalovirus UL112-113 proteins for viral growth and oriLyt-dependent DNA replication.J Microbiol. 2015 Aug;53(8):561-9. doi: 10.1007/s12275-015-5301-3. Epub 2015 Jul 31. J Microbiol. 2015. PMID: 26224459
-
PUL21a-Cyclin A2 interaction is required to protect human cytomegalovirus-infected cells from the deleterious consequences of mitotic entry.PLoS Pathog. 2014 Nov 13;10(10):e1004514. doi: 10.1371/journal.ppat.1004514. eCollection 2014 Oct. PLoS Pathog. 2014. PMID: 25393019 Free PMC article.
-
Novel immediate-early protein IE19 of human cytomegalovirus activates the origin recognition complex I promoter in a cooperative manner with IE72.J Virol. 2002 Apr;76(7):3158-67. doi: 10.1128/jvi.76.7.3158-3167.2002. J Virol. 2002. PMID: 11884540 Free PMC article.
-
The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner.PLoS One. 2012;7(11):e49630. doi: 10.1371/journal.pone.0049630. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166733 Free PMC article.
-
Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression.J Virol. 2001 Nov;75(22):10683-95. doi: 10.1128/JVI.75.22.10683-10695.2001. J Virol. 2001. PMID: 11602710 Free PMC article.
References
-
- Ahn J-H, Chiou C-J, Hayward G S. Evaluation and mapping of the DNA binding and dimerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1997;210:25–36. - PubMed
-
- Ahn, J.-H., and G. S. Hayward. Disruption of PML-associated nuclear bodies by IE1 is required for efficient early stages of viral gene expression and replication in human cytomegalovirus infection. Submitted for publication. - PubMed
-
- Ahn, J.-H., and G. S. Hayward. Unpublished data.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

