Allelic exclusion of the T cell receptor beta locus requires the SH2 domain-containing leukocyte protein (SLP)-76 adaptor protein
- PMID: 10523607
- PMCID: PMC2195661
- DOI: 10.1084/jem.190.8.1093
Allelic exclusion of the T cell receptor beta locus requires the SH2 domain-containing leukocyte protein (SLP)-76 adaptor protein
Abstract
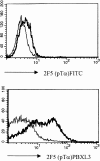
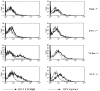
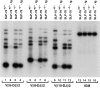
Signaling via the pre-T cell receptor (TCR) is required for the proliferative expansion and maturation of CD4(-)CD8(-) double-negative (DN) thymocytes into CD4(+)CD8(+) double-positive (DP) cells and for TCR-beta allelic exclusion. The adaptor protein SH2 domain-containing leukocyte protein (SLP)-76 has been shown to play a crucial role in thymic development, because thymocytes of SLP-76(-/-) mice are arrested at the CD25(+)CD44(-) DN stage. Here we show that SLP-76(-/-) DN thymocytes express the pre-TCR on their surfaces and that introduction of a TCR-alpha/beta transgene into the SLP-76(-/-) background fails to cause expansion of DN thymocytes or developmental progression to the DP stage. Moreover, analysis of TCR-beta rearrangement in SLP-76(-/-) TCR-transgenic mice or in single CD25(+)CD44(-) DN cells from SLP-76(-/-) mice indicates an essential role of SLP-76 in TCR-beta allelic exclusion.
Figures





Similar articles
-
Conditional deletion reveals a cell-autonomous requirement of SLP-76 for thymocyte selection.J Exp Med. 2005 Oct 3;202(7):893-900. doi: 10.1084/jem.20051128. Epub 2005 Sep 26. J Exp Med. 2005. PMID: 16186188 Free PMC article.
-
Impaired thymic selection in mice expressing altered levels of the SLP-76 adaptor protein.J Leukoc Biol. 2008 Feb;83(2):419-29. doi: 10.1189/jlb.0507297. Epub 2007 Oct 26. J Leukoc Biol. 2008. PMID: 17965338
-
T lymphocyte development in p56lck deficient mice: allelic exclusion of the TcR beta locus is incomplete but thymocyte development is not restored by TcR beta or TcR alpha beta transgenes.Eur J Immunol. 1995 May;25(5):1312-8. doi: 10.1002/eji.1830250527. Eur J Immunol. 1995. PMID: 7774634
-
The role of SLP-76 and LAT in lymphocyte development.Curr Opin Immunol. 2000 Apr;12(2):173-8. doi: 10.1016/s0952-7915(99)00068-0. Curr Opin Immunol. 2000. PMID: 10712938 Review.
-
The Role of Adaptor Proteins in the Biology of Natural Killer T (NKT) Cells.Front Immunol. 2019 Jun 25;10:1449. doi: 10.3389/fimmu.2019.01449. eCollection 2019. Front Immunol. 2019. PMID: 31293596 Free PMC article. Review.
Cited by
-
Pro-B cells sense productive immunoglobulin heavy chain rearrangement irrespective of polypeptide production.Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10644-9. doi: 10.1073/pnas.1019224108. Epub 2011 Jun 13. Proc Natl Acad Sci U S A. 2011. PMID: 21670279 Free PMC article.
-
Conditional deletion reveals a cell-autonomous requirement of SLP-76 for thymocyte selection.J Exp Med. 2005 Oct 3;202(7):893-900. doi: 10.1084/jem.20051128. Epub 2005 Sep 26. J Exp Med. 2005. PMID: 16186188 Free PMC article.
-
Restriction of endogenous T cell antigen receptor beta rearrangements to Vbeta14 through selective recombination signal sequence modifications.Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):4002-7. doi: 10.1073/pnas.0700081104. Epub 2007 Feb 27. Proc Natl Acad Sci U S A. 2007. PMID: 17360467 Free PMC article.
-
Disorderly conduct in gammadelta versus alphabeta T cell lineage commitment.Semin Immunol. 2010 Aug;22(4):222-7. doi: 10.1016/j.smim.2010.04.003. Epub 2010 May 6. Semin Immunol. 2010. PMID: 20451409 Free PMC article. Review.
-
The E delta enhancer controls the generation of CD4- CD8- alphabetaTCR-expressing T cells that can give rise to different lineages of alphabeta T cells.J Exp Med. 2006 Jun 12;203(6):1543-50. doi: 10.1084/jem.20051711. Epub 2006 Jun 5. J Exp Med. 2006. PMID: 16754716 Free PMC article.
References
-
- von Boehmer H., Fehling H.J. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 1997;15:433–452. - PubMed
-
- Fehling H.J., Krotkova A., Saint-Ruf C., von Boehmer H. Crucial role of the pre-T-cell receptor a gene in development of αβ but not γδ T cells. Nature. 1995;375:795–798. - PubMed
-
- Mombaerts P., Clarke A.R., Rudnicki M.A., Iacomini J., Itohara S., Lafaille J.J., Wang L., Ichikawa Y., Jaenisch R., Hooper M.L. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature. 1992;360:225–231. - PubMed
-
- Molina T.J., Kishihara K., Siderovski D.P., van Ewijk W., Narendran A., Timms E., Wakeham A., Paige C.J., Hartmann K.-U., Veilette A. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

