SPI-1-dependent host range of rabbitpox virus and complex formation with cathepsin G is associated with serpin motifs
- PMID: 10516006
- PMCID: PMC112932
- DOI: 10.1128/JVI.73.11.8999-9010.1999
SPI-1-dependent host range of rabbitpox virus and complex formation with cathepsin G is associated with serpin motifs
Abstract
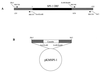
Serpins are a superfamily of serine proteinase inhibitors which function to regulate a number of key biological processes including fibrinolysis, inflammation, and cell migration. Poxviruses are the only viruses known to encode functional serpins. While some poxvirus serpins regulate inflammation (myxoma virus SERP1 and cowpox virus [CPV] crmA/SPI-2) or apoptosis (myxoma virus SERP2 and CPV crmA/SPI-2), the function of other poxvirus serpins remains unknown. The rabbitpox virus (RPV) SPI-1 protein is 47% identical to crmA and shares all of the serpin structural motifs. However, no serpin-like activity has been demonstrated for SPI-1 to date. Earlier we showed that RPV with the SPI-1 gene deleted, unlike wild-type virus, fails to grow on A549 or PK15 cells (A. Ali, P. C. Turner, M. A. Brooks, and R. W. Moyer, Virology 202:306-314, 1994). Here we demonstrate that in the absence of a functional SPI-1 protein, infected nonpermissive cells which exhibit the morphological features of apoptosis fail to activate terminal caspases or cleave the death substrates PARP or lamin A. We show that SPI-1 forms a stable complex in vitro with cathepsin G, a member of the chymotrypsin family of serine proteinases, consistent with serpin activity. SPI-1 reactive-site loop (RSL) mutations of the critical P1 and P14 residues abolish this activity. Viruses containing the SPI-1 RSL P1 or P14 mutations also fail to grow on A549 or PK15 cells. These results suggest that the full virus host range depends on the serpin activity of SPI-1 and that in restrictive cells SPI-1 inhibits a proteinase with chymotrypsin-like activity and may function to inhibit a caspase-independent pathway of apoptosis.
Figures
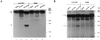






Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article.
-
Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article.
-
Pharmacological treatments in panic disorder in adults: a network meta-analysis.Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
-
Human FAM111A inhibits vaccinia virus replication by degrading viral protein I3 and is antagonized by poxvirus host range factor SPI-1.Proc Natl Acad Sci U S A. 2023 Aug 29;120(35):e2304242120. doi: 10.1073/pnas.2304242120. Epub 2023 Aug 22. Proc Natl Acad Sci U S A. 2023. PMID: 37607234 Free PMC article.
-
Suppressors of a host range mutation in the rabbitpox virus serpin SPI-1 map to proteins essential for viral DNA replication.J Virol. 2005 Jul;79(14):9168-79. doi: 10.1128/JVI.79.14.9168-9179.2005. J Virol. 2005. PMID: 15994811 Free PMC article.
-
Murine serpin 2A is a redox-sensitive intracellular protein.Biochem J. 2003 Apr 1;371(Pt 1):165-73. doi: 10.1042/BJ20021567. Biochem J. 2003. PMID: 12470299 Free PMC article.
-
Mutational analysis of vaccinia virus E3 protein: the biological functions do not correlate with its biochemical capacity to bind double-stranded RNA.J Virol. 2015 May;89(10):5382-94. doi: 10.1128/JVI.03288-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25740987 Free PMC article.
-
Induction of potent humoral and cell-mediated immune responses by attenuated vaccinia virus vectors with deleted serpin genes.J Virol. 2004 Mar;78(6):2770-9. doi: 10.1128/jvi.78.6.2770-2779.2004. J Virol. 2004. PMID: 14990697 Free PMC article.
References
-
- Ali A N, Turner P C, Brooks M A, Moyer R W. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology. 1994;202:306–314. - PubMed
-
- Boyle D B, Coupar B E. A dominant selectable marker for the construction of recombinant poxviruses. Gene. 1988;65:123–128. - PubMed
-
- Bump N J, Hackett M, Hugunin M, Seshagriri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Licari L P P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Inhibition of the ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

