Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA
- PMID: 10500154
- PMCID: PMC18011
- DOI: 10.1073/pnas.96.20.11200
Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA
Abstract
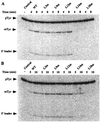
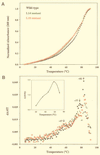
M1 RNA, the catalytic subunit of Escherichia coli RNase P, forms a secondary structure that includes five sequence variants of the tetraloop motif. Site-directed mutagenesis of the five tetraloops of M1 RNA, and subsequent steady-state kinetic analysis in vitro, with different substrates in the presence and absence of the protein cofactor, reveal that (i) certain mutants exhibit defects that vary in a substrate-dependent manner, and that (ii) the protein cofactor can correct the mutant phenotypes in vitro, a phenomenon that is also substrate dependent. Thermal denaturation curves of tetraloop mutants that exhibit kinetic defects differ from those of wild-type M1 RNA. Although the data collected in vitro underscore the importance of the tetraloop motif to M1 RNA function and structure, three of the five tetraloops we examined in vivo are essential for the function of E. coli RNase P. The kinetic data in vitro are not in total agreement with previous phylogenetic predictions but the data in vivo are, as only mutants in those tetraloops proposed to be involved in tertiary interactions fail to complement in vivo. Therefore, the tetraloop motif is critical for the stabilization of the structure of M1 RNA and essential to RNase P function in the cell.
Figures

Similar articles
-
Contributions of phylogenetically variable structural elements to the function of the ribozyme ribonuclease P.Biochemistry. 1992 Jan 21;31(2):328-33. doi: 10.1021/bi00117a003. Biochemistry. 1992. PMID: 1370627
-
A novel tertiary interaction in M1 RNA, the catalytic subunit of Escherichia coli RNase P.Nucleic Acids Res. 1993 Aug 25;21(17):3927-33. doi: 10.1093/nar/21.17.3927. Nucleic Acids Res. 1993. PMID: 7690469 Free PMC article.
-
Acquisition of novel catalytic activity by the M1 RNA ribozyme: the cost of molecular adaptation.J Mol Biol. 1999 Oct 1;292(4):931-44. doi: 10.1006/jmbi.1999.3098. J Mol Biol. 1999. PMID: 10525416
-
The first phytoplasma RNase P RNA provides new insights into the sequence requirements of this ribozyme.Nucleic Acids Res. 2001 Jun 15;29(12):2661-5. doi: 10.1093/nar/29.12.2661. Nucleic Acids Res. 2001. PMID: 11410676 Free PMC article.
-
RNase P: variations and uses.J Biol Chem. 2002 Mar 1;277(9):6759-62. doi: 10.1074/jbc.R100067200. Epub 2001 Dec 10. J Biol Chem. 2002. PMID: 11741968 Review. No abstract available.
Cited by
-
Protein cofactors and substrate influence Mg2+-dependent structural changes in the catalytic RNA of archaeal RNase P.Nucleic Acids Res. 2021 Sep 20;49(16):9444-9458. doi: 10.1093/nar/gkab655. Nucleic Acids Res. 2021. PMID: 34387688 Free PMC article.
-
The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions.Biomolecules. 2016 May 13;6(2):27. doi: 10.3390/biom6020027. Biomolecules. 2016. PMID: 27187488 Free PMC article. Review.
-
Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes.Annu Rev Biochem. 2002;71:165-89. doi: 10.1146/annurev.biochem.71.110601.135352. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045094 Free PMC article. Review.
-
Structural basis of a ribozyme's thermostability: P1-L9 interdomain interaction in RNase P RNA.RNA. 2008 Jan;14(1):127-33. doi: 10.1261/rna.762508. Epub 2007 Nov 12. RNA. 2008. PMID: 17998289 Free PMC article.
-
A subunit of human nuclear RNase P has ATPase activity.Proc Natl Acad Sci U S A. 2001 Jan 16;98(2):441-4. doi: 10.1073/pnas.98.2.441. Epub 2001 Jan 9. Proc Natl Acad Sci U S A. 2001. PMID: 11149958 Free PMC article.
References
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. - PubMed
-
- Altman S, Kirsebom L. In: The RNA World. Gesteland R F, Cech T R, Atkins J P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 351–380.
-
- Cheong C, Varani G, Tinoco I., Jr Nature (London) 1990;346:680–682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

