ATP-Dependent inactivation and sequestration of ornithine decarboxylase by the 26S proteasome are prerequisites for degradation
- PMID: 10490656
- PMCID: PMC84714
- DOI: 10.1128/MCB.19.10.7216
ATP-Dependent inactivation and sequestration of ornithine decarboxylase by the 26S proteasome are prerequisites for degradation
Abstract
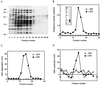
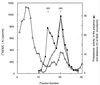
The 26S proteasome is a eukaryotic ATP-dependent protease, but the molecular basis of its energy requirement is largely unknown. Ornithine decarboxylase (ODC) is the only known enzyme to be degraded by the 26S proteasome without ubiquitinylation. We report here that the 26S proteasome is responsible for the irreversible inactivation coupled to sequestration of ODC, a process requiring ATP and antizyme (AZ) but not proteolytic activity. Neither the 20S proteasome (catalytic core) nor PA700 (the regulatory complex) by itself contributed to this ODC inactivation. Analysis with a C-terminal mutant ODC revealed that the 26S proteasome recognizes the C-terminal degradation signal of ODC exposed by attachment of AZ, and subsequent ATP-dependent sequestration of ODC in the 26S proteasome causes irreversible inactivation, possibly unfolding, of ODC and dissociation of AZ. These processes may be linked to the translocation of ODC into the 20S proteasomal inner cavity, centralized within the 26S proteasome, for degradation.
Figures
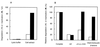



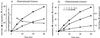


Similar articles
-
Degradation of ornithine decarboxylase by the 26S proteasome.Biochem Biophys Res Commun. 2000 Jan 7;267(1):1-6. doi: 10.1006/bbrc.1999.1706. Biochem Biophys Res Commun. 2000. PMID: 10623564 Review.
-
Degradation of ornithine decarboxylase by the mammalian and yeast 26S proteasome complexes requires all the components of the protease.Eur J Biochem. 1995 Apr 1;229(1):276-83. Eur J Biochem. 1995. PMID: 7744041
-
Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination.Nature. 1992 Dec 10;360(6404):597-9. doi: 10.1038/360597a0. Nature. 1992. PMID: 1334232
-
The 26S proteasome degrades mouse and yeast ornithine decarboxylase in yeast cells.FEBS Lett. 1994 Dec 19;356(2-3):162-4. doi: 10.1016/0014-5793(94)01260-1. FEBS Lett. 1994. PMID: 7805829
-
Antizyme, a mediator of ubiquitin-independent proteasomal degradation.Biochimie. 2001 Mar-Apr;83(3-4):319-23. doi: 10.1016/s0300-9084(01)01252-4. Biochimie. 2001. PMID: 11295492 Review.
Cited by
-
Overproduction of cardiac S-adenosylmethionine decarboxylase in transgenic mice.Biochem J. 2006 Jan 1;393(Pt 1):295-302. doi: 10.1042/BJ20051196. Biochem J. 2006. PMID: 16153183 Free PMC article.
-
A nonproteolytic function of the proteasome is required for the dissociation of Cdc2 and cyclin B at the end of M phase.Genes Dev. 2000 Sep 15;14(18):2344-57. doi: 10.1101/gad.823200. Genes Dev. 2000. PMID: 10995390 Free PMC article.
-
Proteasome substrate degradation requires association plus extended peptide.EMBO J. 2007 Jan 10;26(1):123-31. doi: 10.1038/sj.emboj.7601476. Epub 2006 Dec 7. EMBO J. 2007. PMID: 17170706 Free PMC article.
-
Bifunctional anti-huntingtin proteasome-directed intrabodies mediate efficient degradation of mutant huntingtin exon 1 protein fragments.PLoS One. 2011;6(12):e29199. doi: 10.1371/journal.pone.0029199. Epub 2011 Dec 22. PLoS One. 2011. PMID: 22216210 Free PMC article.
-
Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit.Nucleic Acids Res. 2000 Sep 1;28(17):3185-96. doi: 10.1093/nar/28.17.3185. Nucleic Acids Res. 2000. PMID: 10954585 Free PMC article. Review.
References
-
- Armon T, Ganoth D, Hershko A. Assembly of the 26S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990;265:20723–20726. - PubMed
-
- Auvinen M, Paasinen A, Andersson L C, Hölttä E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–359. - PubMed
-
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. - PubMed
-
- Bercovich Z, Kahana C. Involvement of the 20S proteasome in the degradation of ornithine decarboxylase. Eur J Biochem. 1993;213:205–210. - PubMed
-
- Chu-Ping M, Vu J H, Proske R J, Slaughter C A, DeMartino G N. Identification, purification, and characterization of a high molecular weight ATP-dependent activator (PA700) of the 20S proteasome. J Biol Chem. 1994;269:3539–3547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
