Regulation of RelA subcellular localization by a putative nuclear export signal and p50
- PMID: 10490645
- PMCID: PMC84703
- DOI: 10.1128/MCB.19.10.7088
Regulation of RelA subcellular localization by a putative nuclear export signal and p50
Abstract
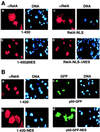
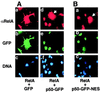
Nuclear factor kappaB (NF-kappaB) represents a family of dimeric DNA binding proteins, the pleotropic form of which is a heterodimer composed of RelA and p50 subunits. The biological activity of NF-kappaB is controlled through its subcellular localization. Inactive NF-kappaB is sequestered in the cytoplasm by physical interaction with an inhibitor, IkappaBalpha. Signal-mediated IkappaBalpha degradation triggers the release and subsequent nuclear translocation of NF-kappaB. It remains unknown whether the NF-kappaB shuttling between the cytoplasm and nucleus is subjected to additional steps of regulation. In this study, we demonstrated that the RelA subunit of NF-kappaB exhibits strong cytoplasmic localization activity even in the absence of IkappaBalpha inhibition. The cytoplasmic distribution of RelA is largely mediated by a leucine-rich sequence homologous to the recently characterized nuclear export signal (NES). This putative NES is both required and sufficient to mediate cytoplasmic localization of RelA as well as that of heterologous proteins. Furthermore, the cytoplasmic distribution of RelA is sensitive to a nuclear export inhibitor, leptomycin B, suggesting that RelA undergoes continuous nuclear export. Interestingly, expression of p50 prevents the cytoplasmic expression of RelA, leading to the nuclear accumulation of both RelA and p50. Together, these results suggest that the nuclear and cytoplasmic shuttling of RelA is regulated by both an intrinsic NES-like sequence and the p50 subunit of NF-kappaB.
Figures







Similar articles
-
A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes.Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):1014-9. doi: 10.1073/pnas.97.3.1014. Proc Natl Acad Sci U S A. 2000. PMID: 10655476 Free PMC article.
-
An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IkappaBalpha.EMBO J. 1999 Dec 1;18(23):6682-93. doi: 10.1093/emboj/18.23.6682. EMBO J. 1999. PMID: 10581242 Free PMC article.
-
Oncoprotein p28 GANK binds to RelA and retains NF-kappaB in the cytoplasm through nuclear export.Cell Res. 2007 Dec;17(12):1020-9. doi: 10.1038/cr.2007.99. Cell Res. 2007. PMID: 18040287
-
Multiple redox regulation in NF-kappaB transcription factor activation.Biol Chem. 1997 Nov;378(11):1237-45. Biol Chem. 1997. PMID: 9426183 Review.
-
Regulation of NF-kappaB action by reversible acetylation.Novartis Found Symp. 2004;259:208-17; discussion 218-25. Novartis Found Symp. 2004. PMID: 15171256 Review.
Cited by
-
Cell-specific association and shuttling of IkappaBalpha provides a mechanism for nuclear NF-kappaB in B lymphocytes.Mol Cell Biol. 2001 Jul;21(14):4837-46. doi: 10.1128/MCB.21.14.4837-4846.2001. Mol Cell Biol. 2001. PMID: 11416157 Free PMC article.
-
Nuclear export of the NF-κB inhibitor IκBα is required for proper B cell and secondary lymphoid tissue formation.Immunity. 2011 Feb 25;34(2):188-200. doi: 10.1016/j.immuni.2011.01.014. Epub 2011 Feb 17. Immunity. 2011. PMID: 21333553 Free PMC article.
-
APEX3 - An Optimized Tool for Rapid and Unbiased Proximity Labeling.J Mol Biol. 2023 Jul 1;435(13):168145. doi: 10.1016/j.jmb.2023.168145. Epub 2023 May 13. J Mol Biol. 2023. PMID: 37182813 Free PMC article.
-
Elements of transcriptional machinery are compatible among plants and mammals.PLoS One. 2013;8(1):e53737. doi: 10.1371/journal.pone.0053737. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326494 Free PMC article.
-
Identification of CRM1-dependent Nuclear Export Cargos Using Quantitative Mass Spectrometry.Mol Cell Proteomics. 2013 Mar;12(3):664-78. doi: 10.1074/mcp.M112.024877. Epub 2012 Dec 13. Mol Cell Proteomics. 2013. PMID: 23242554 Free PMC article.
References
-
- Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. - PubMed
-
- Baeuerle P, Baichwal V R. NF-κB as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv Immunol. 1997;65:111–137. - PubMed
-
- Baeuerle P A. IκB-NF-κB structures: at the interface of inflammation control. Cell. 1998;95:729–731. - PubMed
-
- Baeuerle P A, Baltimore D. A 65-kD subunit of active NF-κB is required for inhibition of NF-κB by IκB. Genes Dev. 1989;3:1689–1698. - PubMed
-
- Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
