The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae
- PMID: 10490601
- PMCID: PMC84637
- DOI: 10.1128/MCB.19.10.6621
The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae
Abstract
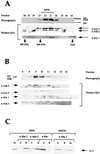
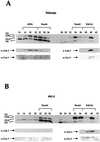
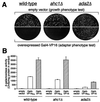
We have identified two Gcn5-dependent histone acetyltransferase (HAT) complexes from Saccharomyces cerevisiae, the 0.8-MDa ADA complex and the 1.8-MDa SAGA complex. The SAGA (Spt-Ada-Gcn5-acetyltransferase) complex contains several subunits which also function as part of other protein complexes, including a subset of TATA box binding protein-associated factors (TAFIIs) and Tra1. These observations raise the question of whether the 0.8-MDa ADA complex is a subcomplex of SAGA or whether it is a distinct HAT complex that also shares subunits with SAGA. To address this issue, we sought to determine if the ADA complex contained subunits that are not present in the SAGA complex. In this study, we report the purification of the ADA complex over 10 chromatographic steps. By a combination of mass spectrometry analysis and immunoblotting, we demonstrate that the adapter proteins Ada2, Ada3, and Gcn5 are indeed integral components of ADA. Furthermore, we identify the product of the S. cerevisiae gene YOR023C as a novel subunit of the ADA complex and name it Ahc1 for ADA HAT complex component 1. Biochemical functions of YOR023C have not been reported. However, AHC1 in high copy numbers suppresses the cold sensitivity caused by particular mutations in HTA1 (I. Pinto and F. Winston, personal communication), which encodes histone H2A (J. N. Hirschhorn et al., Mol. Cell. Biol. 15:1999-2009, 1995). Deletion of AHC1 disrupted the integrity of the ADA complex but did not affect SAGA or give rise to classic Ada(-) phenotypes. These results indicate that Gcn5, Ada2, and Ada3 function as part of a unique HAT complex (ADA) and represent shared subunits between this complex and SAGA.
Figures




Similar articles
-
The SAGA HAT module is tethered by its SWIRM domain and modulates activity of the SAGA DUB module.Biochim Biophys Acta Gene Regul Mech. 2023 Jun;1866(2):194929. doi: 10.1016/j.bbagrm.2023.194929. Epub 2023 Mar 24. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36965704 Free PMC article.
-
Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex.Genes Dev. 1997 Jul 1;11(13):1640-50. doi: 10.1101/gad.11.13.1640. Genes Dev. 1997. PMID: 9224714
-
Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1.Oncogene. 2002 Feb 21;21(9):1411-22. doi: 10.1038/sj.onc.1205201. Oncogene. 2002. PMID: 11857084
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
-
The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194629. doi: 10.1016/j.bbagrm.2020.194629. Epub 2020 Sep 2. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32890768 Free PMC article. Review.
Cited by
-
The SAGA HAT module is tethered by its SWIRM domain and modulates activity of the SAGA DUB module.Biochim Biophys Acta Gene Regul Mech. 2023 Jun;1866(2):194929. doi: 10.1016/j.bbagrm.2023.194929. Epub 2023 Mar 24. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36965704 Free PMC article.
-
Acetylation of histones and transcription-related factors.Microbiol Mol Biol Rev. 2000 Jun;64(2):435-59. doi: 10.1128/MMBR.64.2.435-459.2000. Microbiol Mol Biol Rev. 2000. PMID: 10839822 Free PMC article. Review.
-
Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis.Plant Mol Biol. 2010 Sep;74(1-2):183-200. doi: 10.1007/s11103-010-9665-9. Epub 2010 Jul 27. Plant Mol Biol. 2010. PMID: 20661629
-
X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity.Genetics. 2005 Jan;169(1):51-63. doi: 10.1534/genetics.104.028613. Epub 2004 Sep 15. Genetics. 2005. PMID: 15371366 Free PMC article.
-
Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin.Mol Cell Biol. 2006 Apr;26(8):3018-28. doi: 10.1128/MCB.26.8.3018-3028.2006. Mol Cell Biol. 2006. PMID: 16581777 Free PMC article.
References
-
- Alland L, Muhle R, Hou H J, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
-
- Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Reigier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. - PubMed
-
- Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
