A nuclear action of the eukaryotic cochaperone RAP46 in downregulation of glucocorticoid receptor activity
- PMID: 10477749
- PMCID: PMC2169481
- DOI: 10.1083/jcb.146.5.929
A nuclear action of the eukaryotic cochaperone RAP46 in downregulation of glucocorticoid receptor activity
Abstract
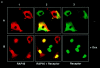
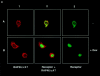
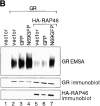
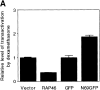
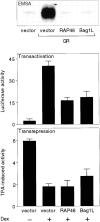
RAP46 is a eukaryotic cochaperone that associates with several proteins, including the heat shock protein hsp70/hsc70 and the glucocorticoid receptor (GR). Here we show a downregulation of GR-mediated transactivation by RAP46 via a mechanism independent of a cytoplasmic action of this cochaperone. We demonstrate a specific cytoplasmic-nuclear recruitment of RAP46 by the liganded GR that results in inhibition of the transactivation function of the receptor. A repeated sequence motif [EEX(4)](8) at the NH(2) terminus of RAP46 or BAG-1L, a larger isoform of RAP46, is responsible for this downregulation of GR activity. BAG-1, a shorter isoform with only a duplication of the [EEX(4)] sequence, does not inhibit GR activity. The [EEX(4)](8) motif, when linked to an otherwise unrelated protein, abrogated the inhibitory action of endogenous RAP46 on GR-mediated transactivation. The nuclear effects of RAP46 and BAG-1L are specific since GR-mediated inhibition of AP-1 activity was not affected. These studies identify the [EEX(4)](8) sequence as a signature motif for inhibition of GR-mediated transactivation and demonstrate a specific nuclear action of a eukaryotic cochaperone in the regulation of GR activity.
Figures




Similar articles
-
Hsp70-RAP46 interaction in downregulation of DNA binding by glucocorticoid receptor.EMBO J. 2000 Dec 1;19(23):6508-16. doi: 10.1093/emboj/19.23.6508. EMBO J. 2000. PMID: 11101523 Free PMC article.
-
BAG-1M: a potential specificity determinant of corticosteroid receptor action.Kidney Int. 2000 Apr;57(4):1265-9. doi: 10.1046/j.1523-1755.2000.00960.x. Kidney Int. 2000. PMID: 10760052 Review.
-
BAG-1 family of cochaperones in the modulation of nuclear receptor action.J Steroid Biochem Mol Biol. 2001 Nov;78(5):379-88. doi: 10.1016/s0960-0760(01)00114-5. J Steroid Biochem Mol Biol. 2001. PMID: 11738548 Review.
-
Bag-1M is a component of the in vivo DNA-glucocorticoid receptor complex at hormone-regulated promoter.J Mol Biol. 2008 Dec 5;384(1):22-30. doi: 10.1016/j.jmb.2008.09.010. Epub 2008 Sep 16. J Mol Biol. 2008. PMID: 18822299
-
RAP46 is a negative regulator of glucocorticoid receptor action and hormone-induced apoptosis.J Biol Chem. 1998 Jun 5;273(23):14620-5. doi: 10.1074/jbc.273.23.14620. J Biol Chem. 1998. PMID: 9603979
Cited by
-
Expression profile of Bag 1 in the postmortem brain.J Chem Neuroanat. 2006 Dec;32(2-4):191-5. doi: 10.1016/j.jchemneu.2006.09.003. Epub 2006 Oct 13. J Chem Neuroanat. 2006. PMID: 17046197 Free PMC article.
-
Mood stabilizer treatment increases serotonin type 1A receptor binding in bipolar depression.J Psychopharmacol. 2013 Oct;27(10):894-902. doi: 10.1177/0269881113499204. Epub 2013 Aug 7. J Psychopharmacol. 2013. PMID: 23926239 Free PMC article. Clinical Trial.
-
Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness.Sci Rep. 2013 Dec 18;3:3539. doi: 10.1038/srep03539. Sci Rep. 2013. PMID: 24345775 Free PMC article.
-
Multiple, but concerted cellular activities of the human protein Hap46/BAG-1M and isoforms.Int J Mol Sci. 2009 Mar;10(3):906-928. doi: 10.3390/ijms10030906. Epub 2009 Mar 2. Int J Mol Sci. 2009. PMID: 19399228 Free PMC article. Review.
-
Cellular mechanisms underlying affective resiliency: the role of glucocorticoid receptor- and mitochondrially-mediated plasticity.Brain Res. 2009 Oct 13;1293:76-84. doi: 10.1016/j.brainres.2009.06.103. Epub 2009 Jul 10. Brain Res. 2009. PMID: 19595676 Free PMC article. Review.
References
-
- Arriza J.L. Aldosterone actionperspectives from the cloning of the mineralocorticoid receptor. In: Fundamental Aspects J.P., Bonvalet N., Farmann M., Lombès, Rafestin-Oblin M.-E., editors. Aldosterone. John Libbey Eurotext Ltd; Paris: 1991. pp. 13–21.
-
- Arriza J.L., Weinberger C., Cerelli G., Glaser T.M., Hendelin B.L., Housmann D.E., Evans R.M. Cloning of human mineralocorticoid receptor complementary DNAstructural and functional kinship with the glucocorticoid receptor. Science. 1987;237:268–273. - PubMed
-
- Beato M., Herrlich P., Schütz G. Steroid hormone receptorsmany actors in search of a plot. Cell. 1995;83:851–857. - PubMed
-
- Bruner K.L., Derfoul A., Robertson N.M., Guerriero G., Fernandes-Alnemri T., Alnemri E.S., Litwack G. The unliganded mineralocorticoid receptor is associated with heat shock proteins 70 and 90 and the immunophilin FKBP-52. Recept. Signal Transduct. 1997;7:85–98. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

