Analysis of BvgA activation of the pertactin gene promoter in Bordetella pertussis
- PMID: 10464192
- PMCID: PMC94027
- DOI: 10.1128/JB.181.17.5234-5241.1999
Analysis of BvgA activation of the pertactin gene promoter in Bordetella pertussis
Abstract
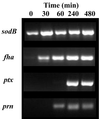
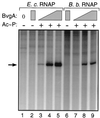
Bordetella pertussis, the causative agent of whooping cough, regulates expression of its virulence factors via a two-component signal transduction system encoded by the bvg regulatory locus. It has been shown by activation kinetics that several of the virulence factors are differentially regulated. fha is transcribed at 10 min following an inducing signal, while ptx is not transcribed until 2 to 4 h after the inducing signal. We present data indicating that prn is transcribed at 1 h, an intermediate time compared to those of fha and ptx. We have identified cis-acting sequences necessary for expression of prn in B. pertussis by using prn-lac fusions containing alterations in the sequence upstream of the prn open reading frame. In vitro transcription and DNase I footprinting analyses provided evidence to support our hypothesis that BvgA binds to this sequence upstream of prn to activate transcription from the promoter. Our genetic data indicate that the region critical for prn activation extends upstream to position -84. However, these data do not support the location of the prn transcription start site as previously published. We used a number of methods, including prn-lac fusions, reverse transcriptase PCR, and 5' rapid amplification of cDNA ends, to localize and identify the bvg-dependent 5' end of the prn transcript to the cytosine at -125 with respect to the published start site.
Figures



Similar articles
-
Differential regulation of Bvg-activated virulence factors plays a role in Bordetella pertussis pathogenicity.Infect Immun. 2001 Apr;69(4):1983-93. doi: 10.1128/IAI.69.4.1983-1993.2001. Infect Immun. 2001. PMID: 11254549 Free PMC article.
-
Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter.J Bacteriol. 1997 Mar;179(5):1755-63. doi: 10.1128/jb.179.5.1755-1763.1997. J Bacteriol. 1997. PMID: 9045838 Free PMC article.
-
Analysis of bvgR expression in Bordetella pertussis.J Bacteriol. 2003 Dec;185(23):6902-12. doi: 10.1128/JB.185.23.6902-6912.2003. J Bacteriol. 2003. PMID: 14617654 Free PMC article.
-
Signal transduction and virulence regulation in Bordetella pertussis.Microbiologia. 1996 Jun;12(2):185-96. Microbiologia. 1996. PMID: 8767703 Review.
-
The Bordetella pertussis model of exquisite gene control by the global transcription factor BvgA.Microbiology (Reading). 2012 Jul;158(Pt 7):1665-1676. doi: 10.1099/mic.0.058941-0. Epub 2012 May 24. Microbiology (Reading). 2012. PMID: 22628479 Free PMC article. Review.
Cited by
-
Differential regulation of Bvg-activated virulence factors plays a role in Bordetella pertussis pathogenicity.Infect Immun. 2001 Apr;69(4):1983-93. doi: 10.1128/IAI.69.4.1983-1993.2001. Infect Immun. 2001. PMID: 11254549 Free PMC article.
-
Multiple weak interactions between BvgA~P and ptx promoter DNA strongly activate transcription of pertussis toxin genes in Bordetella pertussis.PLoS Pathog. 2020 May 13;16(5):e1008500. doi: 10.1371/journal.ppat.1008500. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32401811 Free PMC article.
-
Combined transcriptomic and ChIPseq analyses of the Bordetella pertussis RisA regulon.mSystems. 2024 Apr 16;9(4):e0095123. doi: 10.1128/msystems.00951-23. Epub 2024 Mar 12. mSystems. 2024. PMID: 38470037 Free PMC article.
-
Microarray and functional analysis of growth phase-dependent gene regulation in Bordetella bronchiseptica.Infect Immun. 2009 Oct;77(10):4221-31. doi: 10.1128/IAI.00136-09. Epub 2009 Aug 10. Infect Immun. 2009. PMID: 19667046 Free PMC article.
-
Overexpression of the RNA polymerase alpha subunit reduces transcription of Bvg-activated virulence genes in Bordetella pertussis.J Bacteriol. 2000 Jan;182(2):529-31. doi: 10.1128/JB.182.2.529-531.2000. J Bacteriol. 2000. PMID: 10629205 Free PMC article.
References
-
- Boucher P E, Menozzi F D, Locht C. The modular architecture of bacterial response regulators: insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J Mol Biol. 1994;241:363–377. - PubMed
-
- Carbonetti N H, Irish T J, Chen C H, O’Connell C B, Hadley G A, McNamara U, Tuskan R G, Lewis G K. Intracellular delivery of a cytolytic T-lymphocyte epitope peptide by pertussis toxin to major histocompatability complex class I without involvement of the cytosolic class I antigen processing pathway. Infect Immun. 1999;67:602–607. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

