Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts
- PMID: 10438861
- PMCID: PMC104298
- DOI: 10.1128/JVI.73.9.7710-7721.1999
Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts
Abstract
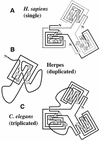
dUTPase is a ubiquitous and essential enzyme responsible for regulating cellular levels of dUTP. The dut gene exists as single, tandemly duplicated, and tandemly triplicated copies. Crystallized single-copy dUTPases have been shown to assemble as homotrimers. dUTPase is encoded as an auxiliary gene in a number of virus genomes. The origin of viral dut genes has remained unresolved since their initial discovery. A comprehensive analysis of dUTPase amino acid sequence relationships was performed to explore the evolutionary dynamics of dut in viruses and their hosts. Our data set, comprised of 24 host and 51 viral sequences, includes representative sequences from available eukaryotes, archaea, eubacteria cells, and viruses, including herpesviruses. These amino acid sequences were aligned by using a hidden Markov model approach developed to align divergent data. Known secondary structures from single-copy crystals were mapped onto the aligned duplicate and triplicate sequences. We show how duplicated dUTPases might fold into a monomer, and we hypothesize that triplicated dUTPases also assemble as monomers. Phylogenetic analysis revealed at least five viral dUTPase sequence lineages in well-supported monophyletic clusters with eukaryotic, eubacterial, and archaeal hosts. We have identified all five as strong examples of horizontal transfer as well as additional potential transfer of dut genes among eubacteria, between eubacteria and viruses, and between retroviruses. The evidence for horizontal transfers is particularly interesting since eukaryotic dut genes have introns, while DNA virus dut genes do not. This implies that an intermediary retroid agent facilitated the horizontal transfer process between host mRNA and DNA viruses.
Figures

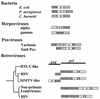
 , the DNA synthesis flavoprotein;
, the DNA synthesis flavoprotein;  , unknown function;
, unknown function;  , the phosphomannomutase;
, the phosphomannomutase;  , the outer membrane protein PIB. Herpesvirus genes:
, the outer membrane protein PIB. Herpesvirus genes:  , the ribonucleotide reductase-related protein;
, the ribonucleotide reductase-related protein;  , the primase. Poxviruses:
, the primase. Poxviruses:  , the transcription initiation factor;
, the transcription initiation factor;  , the ribonucleotide reductase. Retroviruses:
, the ribonucleotide reductase. Retroviruses:  , the protease;
, the protease;  , the reverse transcriptase;
, the reverse transcriptase;  , the ribonuclease H;
, the ribonuclease H;  , the integrase. Unidentified homologous reading frames are indicated (
, the integrase. Unidentified homologous reading frames are indicated ( ). Unidentified nonhomologous reading frames are indicated with a question mark.
). Unidentified nonhomologous reading frames are indicated with a question mark.

Similar articles
-
Evolution of the DUT gene: horizontal transfer between host and pathogen in all three domains of life.Curr Protein Pept Sci. 2001 Dec;2(4):313-24. doi: 10.2174/1389203013381062. Curr Protein Pept Sci. 2001. PMID: 12369928 Review.
-
Biochemical and phylogenetic characterization of the dUTPase from the archaeal virus SIRV.J Biol Chem. 1998 Mar 13;273(11):6024-9. doi: 10.1074/jbc.273.11.6024. J Biol Chem. 1998. PMID: 9497317
-
Potential steps in the evolution of a fused trimeric all-β dUTPase involve a catalytically competent fused dimeric intermediate.FEBS J. 2016 Sep;283(18):3268-86. doi: 10.1111/febs.13800. Epub 2016 Jul 27. FEBS J. 2016. PMID: 27380921
-
Characterization of an early gene encoding for dUTPase in Rana grylio virus.Virus Res. 2007 Feb;123(2):128-37. doi: 10.1016/j.virusres.2006.08.007. Epub 2006 Sep 20. Virus Res. 2007. PMID: 16989917
-
Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review.
Cited by
-
Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.mBio. 2019 Oct 29;10(5):e02483-19. doi: 10.1128/mBio.02483-19. mBio. 2019. PMID: 31662460 Free PMC article.
-
Intra-chain 3D segment swapping spawns the evolution of new multidomain protein architectures.J Mol Biol. 2012 Jan 6;415(1):221-35. doi: 10.1016/j.jmb.2011.10.045. Epub 2011 Nov 4. J Mol Biol. 2012. PMID: 22079367 Free PMC article.
-
The Type 2 dUTPase of Bacteriophage ϕNM1 Initiates Mobilization of Staphylococcus aureus Bovine Pathogenicity Island 1.J Mol Biol. 2016 Jan 16;428(1):142-152. doi: 10.1016/j.jmb.2015.11.009. Epub 2015 Nov 14. J Mol Biol. 2016. PMID: 26585401 Free PMC article.
-
New genes from old: redeployment of dUTPase by herpesviruses.J Virol. 2005 Oct;79(20):12880-92. doi: 10.1128/JVI.79.20.12880-12892.2005. J Virol. 2005. PMID: 16188990 Free PMC article.
-
Phosphorylation of herpes simplex virus 1 dUTPase regulates viral virulence and genome integrity by compensating for low cellular dUTPase activity in the central nervous system.J Virol. 2015 Jan;89(1):241-8. doi: 10.1128/JVI.02497-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320299 Free PMC article.
References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. - PubMed
-
- Caradonna S J, Adamkiewicz D M. Purification and properties of the deoxyuridine triphosphate nucleotidohydrolase enzyme derived from HeLa S3 cells. J Biol Chem. 1984;259:5459–5464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

