Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication
- PMID: 10438837
- PMCID: PMC104274
- DOI: 10.1128/JVI.73.9.7474-7488.1999
Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication
Abstract
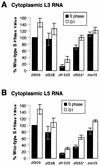
Adenoviruses bearing lesions in the E1B 55-kDa protein (E1B 55-kDa) gene are restricted by the cell cycle such that mutant virus growth is most impaired in cells infected during G(1) and least restricted in cells infected during S phase (F. D. Goodrum and D. A. Ornelles, J. Virol. 71:548-561, 1997). A similar defect is reported here for E4 orf6-mutant viruses. An E4 orf3-mutant virus was not restricted for growth by the cell cycle. However, orf3 was required for enhanced growth of an E4 orf6-mutant virus in cells infected during S phase. The cell cycle restriction may be linked to virus-mediated mRNA transport because both E1B 55-kDa- and E4 orf6-mutant viruses are defective at regulating mRNA transport at late times of infection. Accordingly, the cytoplasmic-to-nuclear ratio of late viral mRNA was reduced in G(1) cells infected with the mutant viruses compared to that in G(1) cells infected with the wild-type virus. By contrast, this ratio was equivalent among cells infected during S phase with the wild-type or mutant viruses. Furthermore, cells infected during S phase with the E1B 55-kDa- or E4 orf6-mutant viruses synthesized more late viral protein than did cells infected during G(1). However, the total amount of cytoplasmic late viral mRNA was greater in cells infected during G(1) than in cells infected during S phase with either the wild-type or mutant viruses, indicating that enhanced transport of viral mRNA in cells infected during S phase cannot account for the difference in yields in cells infected during S phase and in cells infected during G(1). Thus, additional factors affect the cell cycle restriction. These results indicate that the E4 orf6 and orf3 proteins, in addition to the E1B 55-kDa protein, may cooperate to promote cell cycle-independent adenovirus growth.
Figures

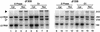
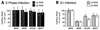
Similar articles
-
Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background.J Virol. 2004 Sep;78(18):9924-35. doi: 10.1128/JVI.78.18.9924-9935.2004. J Virol. 2004. PMID: 15331726 Free PMC article.
-
Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism.J Virol. 2002 May;76(9):4507-19. doi: 10.1128/jvi.76.9.4507-4519.2002. J Virol. 2002. PMID: 11932416 Free PMC article.
-
p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection.J Virol. 1998 Dec;72(12):9479-90. doi: 10.1128/JVI.72.12.9479-9490.1998. J Virol. 1998. PMID: 9811681 Free PMC article.
-
Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins.Curr Top Microbiol Immunol. 2003;272:287-330. doi: 10.1007/978-3-662-05597-7_10. Curr Top Microbiol Immunol. 2003. PMID: 12747554 Review.
-
Genetically modified adenoviruses against gliomas: from bench to bedside.Neurology. 2004 Aug 10;63(3):418-26. doi: 10.1212/01.wnl.0000133302.15022.7f. Neurology. 2004. PMID: 15304571 Review.
Cited by
-
E4orf1 limits the oncolytic potential of the E1B-55K deletion mutant adenovirus.J Virol. 2009 Mar;83(6):2406-16. doi: 10.1128/JVI.01972-08. Epub 2009 Jan 7. J Virol. 2009. PMID: 19129452 Free PMC article.
-
Oncolytic Replication of E1b-Deleted Adenoviruses.Viruses. 2015 Nov 6;7(11):5767-79. doi: 10.3390/v7112905. Viruses. 2015. PMID: 26561828 Free PMC article. Review.
-
Adenovirus replaces mitotic checkpoint controls.J Virol. 2015 May;89(9):5083-96. doi: 10.1128/JVI.00213-15. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694601 Free PMC article.
-
Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription.PLoS Pathog. 2012 Sep;8(9):e1002929. doi: 10.1371/journal.ppat.1002929. Epub 2012 Sep 27. PLoS Pathog. 2012. PMID: 23028327 Free PMC article.
-
Conditionally replicating adenoviruses for cancer treatment.Curr Cancer Drug Targets. 2007 May;7(3):285-301. doi: 10.2174/156800907780618301. Curr Cancer Drug Targets. 2007. PMID: 17504125 Free PMC article. Review.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K S, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and John Wiley and Sons, Inc.; 1993.
-
- Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

