Tandemization of a subregion of the enhancer sequences from SRS 19-6 murine leukemia virus associated with T-lymphoid but not other leukemias
- PMID: 10438804
- PMCID: PMC104241
- DOI: 10.1128/JVI.73.9.7175-7184.1999
Tandemization of a subregion of the enhancer sequences from SRS 19-6 murine leukemia virus associated with T-lymphoid but not other leukemias
Abstract
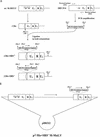
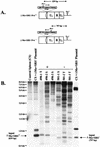
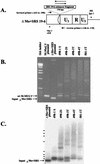
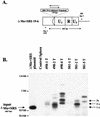
Most simple retroviruses induce tumors of a single cell type when infected into susceptible hosts. The SRS 19-6 murine leukemia virus (MuLV), which originated in mainland China, induces leukemias of multiple cellular origins. Indeed, infected mice often harbor more than one tumor type. Since the enhancers of many MuLVs are major determinants of tumor specificity, we tested the role of the SRS 19-6 MuLV enhancers in its broad disease specificity. The enhancer elements of the Moloney MuLV (M-MuLV) were replaced by the 170-bp enhancers of SRS 19-6 MuLV, yielding the recombinants DeltaMo+SRS(+) and DeltaMo+SRS(-) M-MuLV. M-MuLV normally induces T-lymphoid tumors in all infected mice. Surprisingly, when neonatal mice were inoculated with DeltaMo+SRS(+) or DeltaMo+SRS(-) M-MuLV, all tumors were of T-lymphoid origin, typical of M-MuLV rather than SRS 19-6 MuLV. Thus, the SRS 19-6 MuLV enhancers did not confer the broad disease specificity of SRS 19-6 MuLV to M-MuLV. However, all tumors contained DeltaMo+SRS M-MuLV proviruses with common enhancer alterations. These alterations consisted of tandem multimerization of a subregion of the SRS 19-6 enhancers, encompassing the conserved LVb and core sites and adjacent sequences. Moreover, when tumors induced by the parental SRS 19-6 MuLV were analyzed, most of the T-lymphoid tumors had similar enhancer alterations in the same region whereas tumors of other lineages retained the parental SRS 19-6 MuLV enhancers. These results emphasize the importance of a subregion of the SRS 19-6 MuLV enhancer in induction of T-cell lymphoma. The relevant sequences were consistent with crucial sequences for T-cell lymphomagenesis identified for other MuLVs such as M-MuLV and SL3-3 MuLV. These results also suggest that other regions of the SRS 19-6 MuLV genome contribute to its broad leukemogenic spectrum.
Figures



Similar articles
-
Chimeras between SRS and Moloney murine leukemia viruses reveal novel determinants in disease specificity and MCF recombinant formation.Virology. 2006 Jul 20;351(1):7-17. doi: 10.1016/j.virol.2006.03.010. Epub 2006 Apr 17. Virology. 2006. PMID: 16616947
-
Addition of substitution of simian virus 40 enhancer sequences into the Moloney murine leukemia virus (M-MuLV) long terminal repeat yields infectious M-MuLV with altered biological properties.J Virol. 1988 Jul;62(7):2427-36. doi: 10.1128/JVI.62.7.2427-2436.1988. J Virol. 1988. PMID: 2836623 Free PMC article.
-
Leukemogenicity of Moloney murine leukemia viruses carrying polyoma enhancer sequences in the long terminal repeat is dependent on the nature of the inserted polyoma sequences.Virology. 1988 Sep;166(1):58-65. doi: 10.1016/0042-6822(88)90146-8. Virology. 1988. PMID: 2842957
-
Deregulation of enhancer structure, function, and dynamics in acute lymphoblastic leukemia.Trends Immunol. 2021 May;42(5):418-431. doi: 10.1016/j.it.2021.03.005. Epub 2021 Apr 12. Trends Immunol. 2021. PMID: 33858773 Free PMC article. Review.
-
Enhancing B-Cell Malignancies-On Repurposing Enhancer Activity towards Cancer.Cancers (Basel). 2021 Jun 29;13(13):3270. doi: 10.3390/cancers13133270. Cancers (Basel). 2021. PMID: 34210001 Free PMC article. Review.
Cited by
-
Mutation of all Runx (AML1/core) sites in the enhancer of T-lymphomagenic SL3-3 murine leukemia virus unmasks a significant potential for myeloid leukemia induction and favors enhancer evolution toward induction of other disease patterns.J Virol. 2004 Dec;78(23):13216-31. doi: 10.1128/JVI.78.23.13216-13231.2004. J Virol. 2004. PMID: 15542674 Free PMC article.
-
Novel insights into the pathogenesis of the Graffi murine leukemia retrovirus.J Virol. 2006 Apr;80(8):4026-37. doi: 10.1128/JVI.80.8.4026-4037.2006. J Virol. 2006. PMID: 16571819 Free PMC article.
-
Control of pathogenicity and disease specificity of a T-lymphomagenic gammaretrovirus by E-box motifs but not by an overlapping glucocorticoid response element.J Virol. 2009 Jan;83(1):336-46. doi: 10.1128/JVI.01368-08. Epub 2008 Oct 22. J Virol. 2009. PMID: 18945767 Free PMC article.
-
Enhancers in T Cell development and malignant lesions.Cell Death Discov. 2024 Sep 17;10(1):406. doi: 10.1038/s41420-024-02160-7. Cell Death Discov. 2024. PMID: 39284807 Free PMC article. Review.
References
-
- Brightman B K, Chandy K G, Spencer R H, Gupta S, Pattengale P K, Fan H. Characterization of lymphoid tumors induced by a recombinant murine retrovirus carrying the avian v-myc oncogene. Identification of novel (B-lymphoid) tumors in the thymus. J Immunol. 1988;141:2844–2854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

