An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria
- PMID: 10438738
- PMCID: PMC93955
- DOI: 10.1128/JB.181.16.4734-4740.1999
An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria
Abstract
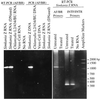
Chlamydia was the only genus in the order Chlamydiales until the recent characterization of Simkania negevensis Z(T) and Parachlamydia acanthamoebae strains. The present study of Chlamydiales 23S ribosomal DNA (rDNA) focuses on a naturally occurring group I intron in the I-CpaI target site of 23S rDNA from S. negevensis. The intron, SnLSU. 1, belonged to the IB4 structural subgroup and was most closely related to large ribosomal subunit introns that express single-motif, LAGLIDADG endonucleases in chloroplasts of algae and in mitochondria of amoebae. RT-PCR and electrophoresis of in vivo rRNA indicated that the intron was not spliced out of the 23S rRNA. The unspliced 658-nt intron is the first group I intron to be found in bacterial rDNA or rRNA, and it may delay the S. negevensis developmental replication cycle by affecting ribosomal function.
Figures



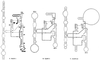

Similar articles
-
Active self-splicing group I introns in 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles.Proc Natl Acad Sci U S A. 2003 Sep 16;100(19):10806-11. doi: 10.1073/pnas.1434268100. Epub 2003 Aug 28. Proc Natl Acad Sci U S A. 2003. PMID: 12947037 Free PMC article.
-
Evolutionary transfer of ORF-containing group I introns between different subcellular compartments (chloroplast and mitochondrion).Mol Biol Evol. 1995 Jul;12(4):533-45. doi: 10.1093/oxfordjournals.molbev.a040234. Mol Biol Evol. 1995. PMID: 7659010
-
Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms.Int J Syst Bacteriol. 1999 Apr;49 Pt 2:415-40. doi: 10.1099/00207713-49-2-415. Int J Syst Bacteriol. 1999. PMID: 10319462
-
Simkania negevensis, an insight into the biology and clinical importance of a novel member of the Chlamydiales order.Crit Rev Microbiol. 2017 Feb;43(1):62-80. doi: 10.3109/1040841X.2016.1165650. Epub 2016 Oct 27. Crit Rev Microbiol. 2017. PMID: 27786615 Review.
-
Parachlamydiaceae: potential emerging pathogens.Emerg Infect Dis. 2002 Jun;8(6):625-30. doi: 10.3201/eid0806.010210. Emerg Infect Dis. 2002. PMID: 12023921 Free PMC article. Review.
Cited by
-
ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae.J Bacteriol. 2004 Feb;186(3):683-91. doi: 10.1128/JB.186.3.683-691.2004. J Bacteriol. 2004. PMID: 14729693 Free PMC article.
-
The unusual 23S rRNA gene of Coxiella burnetii: two self-splicing group I introns flank a 34-base-pair exon, and one element lacks the canonical omegaG.J Bacteriol. 2007 Sep;189(18):6572-9. doi: 10.1128/JB.00812-07. Epub 2007 Jul 20. J Bacteriol. 2007. PMID: 17644584 Free PMC article.
-
Was the Chlamydial Adaptative Strategy to Tryptophan Starvation an Early Determinant of Plastid Endosymbiosis?Front Cell Infect Microbiol. 2016 Jun 22;6:67. doi: 10.3389/fcimb.2016.00067. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27446814 Free PMC article. Review.
-
Active self-splicing group I introns in 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles.Proc Natl Acad Sci U S A. 2003 Sep 16;100(19):10806-11. doi: 10.1073/pnas.1434268100. Epub 2003 Aug 28. Proc Natl Acad Sci U S A. 2003. PMID: 12947037 Free PMC article.
-
Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria.Proc Natl Acad Sci U S A. 2012 Mar 13;109(11):4203-8. doi: 10.1073/pnas.1120192109. Epub 2012 Feb 27. Proc Natl Acad Sci U S A. 2012. PMID: 22371583 Free PMC article.
References
-
- Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

