Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate
- PMID: 10398670
- PMCID: PMC100243
- DOI: 10.1128/CMR.12.3.367
Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate
Abstract
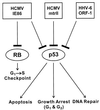
This review is an update on the transforming genes of human cytomegalovirus (HCMV) and human herpesvirus 6 (HHV-6). Both viruses have been implicated in the etiology of several human cancers. In particular, HCMV has been associated with cervical carcinoma and adenocarcinomas of the prostate and colon. In vitro transformation studies have established three HCMV morphologic transforming regions (mtr), i.e., mtrI, mtrII, and mtrIII. Of these, only mtrII (UL111A) is retained and expressed in both transformed and tumor-derived cells. The transforming and tumorigenic activities of the mtrII oncogene were localized to an open reading frame (ORF) encoding a 79-amino-acid (aa) protein. Furthermore, mtrII protein bound to the tumor suppressor protein p53 and inhibited its ability to transactivate a p53-responsive promoter. In additional studies, the HCMV immediate-early protein IE86 (IE2; UL122) was found to interact with cell cycle-regulatory proteins such as p53 and Rb. However, IE86 exhibited transforming activity in vitro only in cooperation with adenovirus E1A. HHV-6 is a T-cell-tropic virus associated with AIDS-related and other lymphoid malignancies. In vitro studies identified three transforming fragments, i.e., SalI-L, ZVB70, and ZVH14. Of these, only SalI-L (DR7) was retained in transformed and tumor-derived cells. The transforming and tumorigenic activities of SalI-L have been localized to a 357-aa ORF-1 protein. The ORF-1 protein was expressed in transformed cells and, like HCMV mtrII, bound to p53 and inhibited its ability to transactivate a p53-responsive promoter. HHV-6 has also been proposed to be a cofactor in AIDS because both HHV-6 and human immunodeficiency virus type 1 (HIV-1) have been demonstrated to coinfect human CD4(+) T cells, causing accelerated cytopathic effects. Interestingly, like the transforming proteins of DNA tumor viruses such as simian virus 40 and adenovirus, ORF-1 was also a transactivator and specifically up-regulated the HIV-1 long terminal repeat when cotransfected into CD4(+) T cells. Finally, based on the interactions of HCMV and HHV-6 transforming proteins with tumor suppressor proteins, a scheme is proposed for their role in oncogenesis.
Figures



Similar articles
-
Human herpesvirus 6 (HHV-6) ORF-1 transactivating gene exhibits malignant transforming activity and its protein binds to p53.Oncogene. 1997 Jan 23;14(3):359-67. doi: 10.1038/sj.onc.1200840. Oncogene. 1997. PMID: 9018122
-
Transcriptional activation of minimal HIV-1 promoter by ORF-1 protein expressed from the SalI-L fragment of human herpesvirus 6.Virology. 1994 May 15;201(1):95-106. doi: 10.1006/viro.1994.1269. Virology. 1994. PMID: 8178493
-
A 79 amino acid oncogene is responsible for human cytomegalovirus mtrII induced malignant transformation.Arch Virol. 1994;136(1-2):161-72. doi: 10.1007/BF01538825. Arch Virol. 1994. PMID: 8002783
-
Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells.Intervirology. 1996;39(4):259-69. doi: 10.1159/000150527. Intervirology. 1996. PMID: 9078467 Review.
-
[Molecular biology of human herpesvirus 6: DNA replication and trans-activator genes].Nihon Rinsho. 1998 Jan;56(1):50-5. Nihon Rinsho. 1998. PMID: 9465664 Review. Japanese.
Cited by
-
Modulation of p53 activity by IkappaBalpha: evidence suggesting a common phylogeny between NF-kappaB and p53 transcription factors.BMC Immunol. 2005 Jun 21;6:12. doi: 10.1186/1471-2172-6-12. BMC Immunol. 2005. PMID: 15969767 Free PMC article.
-
Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10).Proc Natl Acad Sci U S A. 2000 Feb 15;97(4):1695-700. doi: 10.1073/pnas.97.4.1695. Proc Natl Acad Sci U S A. 2000. PMID: 10677520 Free PMC article.
-
β-HHVs and HHV-8 in Lymphoproliferative Disorders.Mediterr J Hematol Infect Dis. 2011;3(1):e2011043. doi: 10.4084/MJHID.2011.043. Epub 2011 Oct 24. Mediterr J Hematol Infect Dis. 2011. PMID: 22110893 Free PMC article.
-
Cytomegalovirus infection may be oncoprotective against neoplasms of B-lymphocyte lineage: single-institution experience and survey of global evidence.Virol J. 2022 Sep 29;19(1):155. doi: 10.1186/s12985-022-01884-1. Virol J. 2022. PMID: 36171605 Free PMC article.
-
Expression of Oncogenic Alleles Induces Multiple Blocks to Human Cytomegalovirus Infection.J Virol. 2016 Apr 14;90(9):4346-4356. doi: 10.1128/JVI.00179-16. Print 2016 May. J Virol. 2016. PMID: 26889030 Free PMC article.
References
-
- Ablashi D V, Josephs S F, Buchbinder A, Hellman K, Nakamura S, Llana T, Lusso P, Kaplan M, Dahlberg J, Memon S, Imam F, Ablashi K L, Markham P D, Kramarsky B, Krueger G R F, Biberfeld P, Wong-Staal F, Salahuddin S Z, Gallo R C. Human B-lymphotropic virus (human herpesvirus-6) J Virol Methods. 1988;21:29–48. - PubMed
-
- Ablashi D V, Salahuddin S Z, Josephs S F, Imam F, Lusso P, Gallo R C, Hung C, Lemp J, Markham P D. HBLV (or HHV-6) in human cell lines. Nature. 1987;329:207. . (Letter.) - PubMed
-
- Albrecht T, Rapp F. Malignant transformation of hamster embryo fibroblasts exposure to ultraviolet-irradiated human cytomegalovirus. Virology. 1973;55:53–61. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Asano Y, Yoshikawa T, Suga S, Yazaki T, Kondo K, Yamanishi K. Fatal fulminant hepatitis in an infant with human herpesvirus-6 infection. Lancet. 1990;335:862–863. . (Letter.) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

