Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences
- PMID: 10373568
- PMCID: PMC84362
- DOI: 10.1128/MCB.19.7.5189
Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences
Abstract
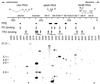
In Drosophila, two classes of genes, the trithorax group and the Polycomb group, are required in concert to maintain gene expression by regulating chromatin structure. We have identified Trithorax protein (TRX) binding elements within the bithorax complex and have found that within the bxd/pbx regulatory region these elements are functionally relevant for normal expression patterns in embryos and confer TRX binding in vivo. TRX was localized to three closely situated sites within a 3-kb chromatin maintenance unit with a modular structure. Results of an in vivo analysis showed that these DNA fragments (each approximately 400 bp) contain both TRX- and Polycomb-group response elements (TREs and PREs) and that in the context of the endogenous Ultrabithorax gene, all of these elements are essential for proper maintenance of expression in embryos. Dissection of one of these maintenance modules showed that TRX- and Polycomb-group responsiveness is conferred by neighboring but separable DNA sequences, suggesting that independent protein complexes are formed at their respective response elements. Furthermore, we have found that the activity of this TRE requires a sequence (approximately 90 bp) which maps to within several tens of base pairs from the closest neighboring PRE and that the PRE activity in one of the elements may require a binding site for PHO, the protein product of the Polycomb-group gene pleiohomeotic. Our results show that long-range maintenance of Ultrabithorax expression requires a complex element composed of cooperating modules, each capable of interacting with both positive and negative chromatin regulators.
Figures
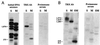
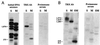

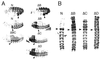


Similar articles
-
Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis.EMBO J. 1998 Sep 1;17(17):5141-50. doi: 10.1093/emboj/17.17.5141. EMBO J. 1998. PMID: 9724650 Free PMC article.
-
Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression.EMBO J. 1997 Jun 16;16(12):3621-32. doi: 10.1093/emboj/16.12.3621. EMBO J. 1997. PMID: 9218803 Free PMC article.
-
GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element.Nucleic Acids Res. 2003 Jul 15;31(14):4147-56. doi: 10.1093/nar/gkg479. Nucleic Acids Res. 2003. PMID: 12853632 Free PMC article.
-
Heritable chromatin states induced by the Polycomb and trithorax group genes.Novartis Found Symp. 1998;214:51-61; discussion 61-6, 104-13. doi: 10.1002/9780470515501.ch4. Novartis Found Symp. 1998. PMID: 9601011 Review.
-
The Polycomb group--no longer an exclusive club?Curr Opin Genet Dev. 2001 Apr;11(2):175-81. doi: 10.1016/s0959-437x(00)00176-3. Curr Opin Genet Dev. 2001. PMID: 11250141 Review.
Cited by
-
Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module.EMBO J. 2004 Feb 25;23(4):857-68. doi: 10.1038/sj.emboj.7600108. Epub 2004 Feb 12. EMBO J. 2004. PMID: 14963490 Free PMC article.
-
Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing.Mol Cell Biol. 2001 Jul;21(14):4528-43. doi: 10.1128/MCB.21.14.4528-4543.2001. Mol Cell Biol. 2001. PMID: 11416132 Free PMC article.
-
Chromatin dynamics: H3K4 methylation and H3 variant replacement during development and in cancer.Cell Mol Life Sci. 2014 Sep;71(18):3439-63. doi: 10.1007/s00018-014-1605-4. Epub 2014 Mar 28. Cell Mol Life Sci. 2014. PMID: 24676717 Free PMC article. Review.
-
batman Interacts with polycomb and trithorax group genes and encodes a BTB/POZ protein that is included in a complex containing GAGA factor.Mol Cell Biol. 2003 Feb;23(4):1181-95. doi: 10.1128/MCB.23.4.1181-1195.2003. Mol Cell Biol. 2003. PMID: 12556479 Free PMC article.
-
Inheritance of Polycomb-dependent chromosomal interactions in Drosophila.Genes Dev. 2003 Oct 1;17(19):2406-20. doi: 10.1101/gad.269503. Genes Dev. 2003. PMID: 14522946 Free PMC article.
References
-
- Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis I B, Hogness D S. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. - PubMed
-
- Bienz M, Müller J. Transcriptional silencing of homeotic genes in Drosophila. BioEssays. 1995;17:775–784. - PubMed
-
- Breen T R, Harte P J. trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. - PubMed
-
- Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
