The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns
- PMID: 10225945
- PMCID: PMC2185073
- DOI: 10.1083/jcb.145.3.425
The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns
Abstract
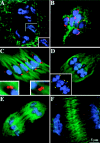
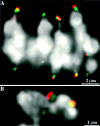
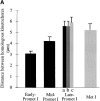
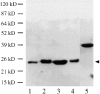
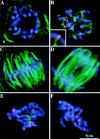
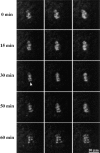
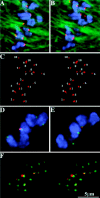
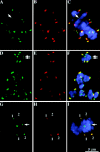
We have identified a maize homologue of yeast MAD2, an essential component in the spindle checkpoint pathway that ensures metaphase is complete before anaphase begins. Combined immunolocalization of MAD2 and a recently cloned maize CENPC homologue indicates that MAD2 localizes to an outer domain of the prometaphase kinetochore. MAD2 staining was primarily observed on mitotic kinetochores that lacked attached microtubules; i.e., at prometaphase or when the microtubules were depolymerized with oryzalin. In contrast, the loss of MAD2 staining in meiosis was not correlated with initial microtubule attachment but was correlated with a measure of tension: the distance between homologous or sister kinetochores (in meiosis I and II, respectively). Further, the tension-sensitive 3F3/2 phosphoepitope colocalized, and was lost concomitantly, with MAD2 staining at the meiotic kinetochore. The mechanism of spindle assembly (discussed here with respect to maize mitosis and meiosis) is likely to affect the relative contributions of attachment and tension. We support the idea that MAD2 is attachment-sensitive and that tension stabilizes microtubule attachments.
Figures


Similar articles
-
Localization of Mad2 to kinetochores depends on microtubule attachment, not tension.J Cell Biol. 1998 Jun 1;141(5):1181-91. doi: 10.1083/jcb.141.5.1181. J Cell Biol. 1998. PMID: 9606210 Free PMC article.
-
Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells.J Cell Biol. 2000 Sep 18;150(6):1233-50. doi: 10.1083/jcb.150.6.1233. J Cell Biol. 2000. PMID: 10995431 Free PMC article.
-
Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells.J Cell Sci. 2004 Apr 1;117(Pt 9):1757-71. doi: 10.1242/jcs.01033. Epub 2004 Mar 16. J Cell Sci. 2004. PMID: 15075237
-
Tension sensors reveal how the kinetochore shares its load.Bioessays. 2017 Jul;39(7):10.1002/bies.201600216. doi: 10.1002/bies.201600216. Epub 2017 Jun 5. Bioessays. 2017. PMID: 28582586 Free PMC article. Review.
-
Merotelic kinetochores in mammalian tissue cells.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):553-68. doi: 10.1098/rstb.2004.1610. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897180 Free PMC article. Review.
Cited by
-
Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension.Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):E1858-67. doi: 10.1073/pnas.1204686109. Epub 2012 May 2. Proc Natl Acad Sci U S A. 2012. PMID: 22552228 Free PMC article.
-
Synchronous nuclear-envelope breakdown and anaphase onset in plant multinucleate cells.Protoplasma. 2001;218(3-4):192-202. doi: 10.1007/BF01306608. Protoplasma. 2001. PMID: 11770435
-
Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner.Mol Cell Biol. 2002 Apr;22(8):2728-42. doi: 10.1128/MCB.22.8.2728-2742.2002. Mol Cell Biol. 2002. PMID: 11909965 Free PMC article.
-
Exploring Plant Meiosis: Insights from the Kinetochore Perspective.Curr Issues Mol Biol. 2023 Sep 28;45(10):7974-7995. doi: 10.3390/cimb45100504. Curr Issues Mol Biol. 2023. PMID: 37886947 Free PMC article. Review.
-
The spindle checkpoint and chromosome segregation in meiosis.FEBS J. 2015 Jul;282(13):2471-87. doi: 10.1111/febs.13166. Epub 2015 Jan 12. FEBS J. 2015. PMID: 25470754 Free PMC article. Review.
References
-
- Anthony RG, Waldin TR, Ray JA, Bright SWJ, Hussey PJ. Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature. 1998;393:260–263. - PubMed
-
- Asai DJ, Brokaw CJ, Thompson WC, Wilson L. Two different monoclonal antibodies to tubulin inhibit the bending of reactivated sea urchin spermatozoa. Cell Motil. 1982;2:599–614. - PubMed
-
- Atschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ault JG, Nicklas RB. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma. 1989;98:33–39. - PubMed
-
- Bajer, A.S., and J. Molé-Bajer. 1972. Spindle Dynamics and Chromosome Movements. Academic Press, New York.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

