Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae
- PMID: 10189369
- PMCID: PMC2148216
- DOI: 10.1083/jcb.145.1.69
Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae
Abstract
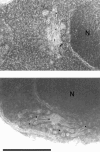
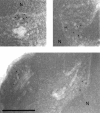
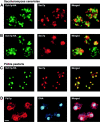
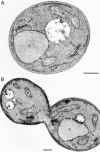
Golgi stacks are often located near sites of "transitional ER" (tER), where COPII transport vesicles are produced. This juxtaposition may indicate that Golgi cisternae form at tER sites. To explore this idea, we examined two budding yeasts: Pichia pastoris, which has coherent Golgi stacks, and Saccharomyces cerevisiae, which has a dispersed Golgi. tER structures in the two yeasts were visualized using fusions between green fluorescent protein and COPII coat proteins. We also determined the localization of Sec12p, an ER membrane protein that initiates the COPII vesicle assembly pathway. In P. pastoris, Golgi stacks are adjacent to discrete tER sites that contain COPII coat proteins as well as Sec12p. This arrangement of the tER-Golgi system is independent of microtubules. In S. cerevisiae, COPII vesicles appear to be present throughout the cytoplasm and Sec12p is distributed throughout the ER, indicating that COPII vesicles bud from the entire ER network. We propose that P. pastoris has discrete tER sites and therefore generates coherent Golgi stacks, whereas S. cerevisiae has a delocalized tER and therefore generates a dispersed Golgi. These findings open the way for a molecular genetic analysis of tER sites.
Figures




Similar articles
-
The transitional ER localization mechanism of Pichia pastoris Sec12.Dev Cell. 2004 May;6(5):649-59. doi: 10.1016/s1534-5807(04)00129-7. Dev Cell. 2004. PMID: 15130490
-
Isolation of Pichia pastoris genes involved in ER-to-Golgi transport.Yeast. 2000 Aug;16(11):979-93. doi: 10.1002/1097-0061(200008)16:11<979::AID-YEA594>3.0.CO;2-C. Yeast. 2000. PMID: 10923020
-
De novo formation of transitional ER sites and Golgi structures in Pichia pastoris.Nat Cell Biol. 2002 Oct;4(10):750-6. doi: 10.1038/ncb852. Nat Cell Biol. 2002. PMID: 12360285
-
The yeast GRASP Grh1 colocalizes with COPII and is dispensable for organizing the secretory pathway.Traffic. 2010 Sep;11(9):1168-79. doi: 10.1111/j.1600-0854.2010.01089.x. Epub 2010 Jun 21. Traffic. 2010. PMID: 20573068 Free PMC article. Review.
-
Exiting the endoplasmic reticulum.Mol Cell Endocrinol. 2001 May 25;177(1-2):13-8. doi: 10.1016/s0303-7207(01)00438-5. Mol Cell Endocrinol. 2001. PMID: 11377815 Review.
Cited by
-
Vesicle-mediated ER export of proteins and lipids.Biochim Biophys Acta. 2012 Aug;1821(8):1040-9. doi: 10.1016/j.bbalip.2012.01.005. Epub 2012 Jan 11. Biochim Biophys Acta. 2012. PMID: 22265716 Free PMC article. Review.
-
Aspergillus nidulans biofilm formation modifies cellular architecture and enables light-activated autophagy.Mol Biol Cell. 2021 Jun 1;32(12):1181-1192. doi: 10.1091/mbc.E20-11-0734. Epub 2021 Apr 7. Mol Biol Cell. 2021. PMID: 33826367 Free PMC article.
-
ER-to-Golgi transport by COPII vesicles in Arabidopsis involves a ribosome-excluding scaffold that is transferred with the vesicles to the Golgi matrix.Protoplasma. 2008 Dec;234(1-4):51-64. doi: 10.1007/s00709-008-0015-6. Epub 2008 Sep 20. Protoplasma. 2008. PMID: 18810574
-
Membrane traffic within the Golgi apparatus.Annu Rev Cell Dev Biol. 2009;25:113-32. doi: 10.1146/annurev.cellbio.24.110707.175421. Annu Rev Cell Dev Biol. 2009. PMID: 19575639 Free PMC article. Review.
-
ER arrival sites for COPI vesicles localize to hotspots of membrane trafficking.EMBO J. 2016 Sep 1;35(17):1935-55. doi: 10.15252/embj.201592873. Epub 2016 Jul 20. EMBO J. 2016. PMID: 27440402 Free PMC article.
References
-
- Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. Signaling via mitogen-activated protein kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. - PubMed
-
- Ayscough K, Hajibagheri NMA, Watson R, Warren G. Stacking of Golgi cisternae in Schizosaccharomyces pomberequires intact microtubules. J Cell Sci. 1993;106:1227–1237. - PubMed
-
- Barlowe C, Schekman R. SEC12encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

