Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis
- PMID: 10074523
- PMCID: PMC88646
- DOI: 10.1128/JCM.37.4.1035-1044.1999
Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis
Abstract
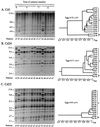
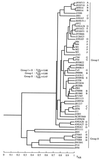
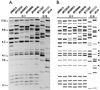
Using a strategy to clone large genomic sequences containing repetitive elements from the infectious yeast Candida dubliniensis, the three unrelated sequences Cd1, Cd24, and Cd25, with respective molecular sizes of 15,500, 10,000, and 16,000 bp, were cloned and analyzed for their efficacy as DNA fingerprinting probes. Each generated a complex Southern blot hybridization pattern with endonuclease-digested genomic DNA. Cd1 generated an extremely variable pattern that contained all of the bands of the pattern generated by the repeat element RPS of Candida albicans. We demonstrated that Cd1 does not contain RPS but does contain a repeat element associated with RPS throughout the C. dubliniensis genome. The Cd1 pattern was the least stable over time both in vitro and in vivo and for that reason proved most effective in assessing microevolution. Cd24, which did not exhibit microevolution in vitro, was highly variable in vivo, suggesting in vivo-dependent microevolution. Cd25 was deemed the best probe for broad epidemiological studies, since it was the most stable over time, was the only truly C. dubliniensis-specific probe of the three, generated the most complex pattern, was distributed throughout all C. dubliniensis chromosomes, and separated a worldwide collection of 57 C. dubliniensis isolates into two distinct groups. The presence of a species-specific repetitive element in Cd25 adds weight to the already substantial evidence that C. dubliniensis represents a bona fide species.
Figures





Similar articles
-
Development and use of complex probes for DNA fingerprinting the infectious fungi.Med Mycol. 2001 Feb;39(1):1-8. doi: 10.1080/mmy.39.1.1.8. Med Mycol. 2001. PMID: 11270395
-
Microevolutionary changes and chromosomal translocations are more frequent at RPS loci in Candida dubliniensis than in Candida albicans.Infect Genet Evol. 2002 Oct;2(1):19-37. doi: 10.1016/s1567-1348(02)00058-8. Infect Genet Evol. 2002. PMID: 12797998
-
Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo.J Clin Microbiol. 2002 Feb;40(2):556-74. doi: 10.1128/JCM.40.2.556-574.2002. J Clin Microbiol. 2002. PMID: 11825972 Free PMC article.
-
Molecular fingerprinting methods for the discrimination between C. albicans and C. dubliniensis.Oral Dis. 2006 May;12(3):242-53. doi: 10.1111/j.1601-0825.2005.01189.x. Oral Dis. 2006. PMID: 16700733 Review.
-
Candida dubliniensis: ten years on.FEMS Microbiol Lett. 2005 Dec 1;253(1):9-17. doi: 10.1016/j.femsle.2005.09.015. Epub 2005 Sep 26. FEMS Microbiol Lett. 2005. PMID: 16213674 Review.
Cited by
-
Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis.Antimicrob Agents Chemother. 2002 Jun;46(6):1695-703. doi: 10.1128/AAC.46.6.1695-1703.2002. Antimicrob Agents Chemother. 2002. PMID: 12019078 Free PMC article.
-
Reduced azole susceptibility in genotype 3 Candida dubliniensis isolates associated with increased CdCDR1 and CdCDR2 expression.Antimicrob Agents Chemother. 2005 Apr;49(4):1312-8. doi: 10.1128/AAC.49.4.1312-1318.2005. Antimicrob Agents Chemother. 2005. PMID: 15793103 Free PMC article.
-
Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel.J Clin Microbiol. 2000 Jan;38(1):170-4. doi: 10.1128/JCM.38.1.170-174.2000. J Clin Microbiol. 2000. PMID: 10618082 Free PMC article.
-
Cross-resistance between fluconazole and ravuconazole and the use of fluconazole as a surrogate marker to predict susceptibility and resistance to ravuconazole among 12,796 clinical isolates of Candida spp.J Clin Microbiol. 2004 Jul;42(7):3137-41. doi: 10.1128/JCM.42.7.3137-3141.2004. J Clin Microbiol. 2004. PMID: 15243072 Free PMC article.
-
Molecular mechanisms of itraconazole resistance in Candida dubliniensis.Antimicrob Agents Chemother. 2003 Aug;47(8):2424-37. doi: 10.1128/AAC.47.8.2424-2437.2003. Antimicrob Agents Chemother. 2003. PMID: 12878500 Free PMC article.
References
-
- Anthony R M, Midgley J, Sweet S P, Howell S A. Multiple strains of Candida albicans in the oral cavity of HIV-positive and HIV-negative patients. Microb Ecol Health Dis. 1995;8:23–30.
-
- Carlotti A, Srikantha T, Schröppel K, Kvaal C, Villard J, Soll D R. A novel repeat sequence (CKRS-1) containing a tandemly repeated subelement (kre) accounts for differences between Candida krusei strains fingerprinted with the probe CkF1,2. Curr Genet. 1997;31:255–263. - PubMed
-
- Chindamporn A, Nakagawa Y, Nizuguchi I, Chibana H, Doi M, Tanaka K. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology. 1998;144:849–857. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

