Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8
- PMID: 10074154
- PMCID: PMC104064
- DOI: 10.1128/JVI.73.4.3040-3053.1999
Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8
Abstract
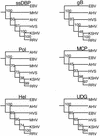
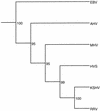
We have sequenced the long unique region (LUR) and characterized the terminal repeats of the genome of a rhesus rhadinovirus (RRV), strain 17577. The LUR as sequenced is 131,364 bp in length, with a G+C content of 52.2% and a CpG ratio of 1.11. The genome codes for 79 open reading frames (ORFs), with 67 of these ORFs similar to genes found in both Kaposi's sarcoma-associated herpesvirus (KSHV) (formal name, human herpesvirus 8) and herpesvirus saimiri. Eight of the 12 unique genes show similarity to genes found in KSHV, including genes for viral interleukin-6, viral macrophage inflammatory protein, and a family of viral interferon regulatory factors (vIRFs). Genomic organization is essentially colinear with KSHV, the primary differences being the number of cytokine and IRF genes and the location of the gene for dihydrofolate reductase. Highly repetitive sequences are located in positions corresponding to repetitive sequences found in KSHV. Phylogenetic analysis of several ORFs supports the similarity between RRV and KSHV. Overall, the sequence, structural, and phylogenetic data combine to provide strong evidence that RRV 17577 is the rhesus macaque homolog of KSHV.
Figures

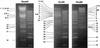




Similar articles
-
The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577.J Virol. 2000 Apr;74(7):3388-98. doi: 10.1128/jvi.74.7.3388-3398.2000. J Virol. 2000. PMID: 10708456 Free PMC article.
-
Complete genome sequence of Pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus.J Virol. 2015 Apr;89(7):3888-909. doi: 10.1128/JVI.03597-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609822 Free PMC article.
-
Next-generation sequence analysis of the genome of RFHVMn, the macaque homolog of Kaposi's sarcoma (KS)-associated herpesvirus, from a KS-like tumor of a pig-tailed macaque.J Virol. 2013 Dec;87(24):13676-93. doi: 10.1128/JVI.02331-13. Epub 2013 Oct 9. J Virol. 2013. PMID: 24109218 Free PMC article.
-
Rhesus monkey rhadinovirus: a model for the study of KSHV.Curr Top Microbiol Immunol. 2007;312:43-69. doi: 10.1007/978-3-540-34344-8_2. Curr Top Microbiol Immunol. 2007. PMID: 17089793 Review.
-
Rhesus macaque rhadinovirus-associated disease.Curr Opin Virol. 2013 Jun;3(3):245-50. doi: 10.1016/j.coviro.2013.05.016. Epub 2013 Jun 6. Curr Opin Virol. 2013. PMID: 23747119 Free PMC article. Review.
Cited by
-
Viral FLICE inhibitory protein of rhesus monkey rhadinovirus inhibits apoptosis by enhancing autophagosome formation.PLoS One. 2012;7(6):e39438. doi: 10.1371/journal.pone.0039438. Epub 2012 Jun 20. PLoS One. 2012. PMID: 22745754 Free PMC article.
-
Viral interleukin-6 encoded by rhesus macaque rhadinovirus is associated with lymphoproliferative disorder (LPD).J Med Primatol. 2009 Oct;38 Suppl 1(Suppl 1):2-7. doi: 10.1111/j.1600-0684.2009.00369.x. J Med Primatol. 2009. PMID: 19863672 Free PMC article.
-
Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses.J Virol. 1999 Dec;73(12):10320-8. doi: 10.1128/JVI.73.12.10320-10328.1999. J Virol. 1999. PMID: 10559350 Free PMC article.
-
Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21(CIP-1).J Virol. 2003 Aug;77(16):8893-914. doi: 10.1128/jvi.77.16.8893-8914.2003. J Virol. 2003. PMID: 12885907 Free PMC article.
-
Transcription of true late (γ2) cytomegalovirus genes requires UL92 function that is conserved among beta- and gammaherpesviruses.J Virol. 2014 Jan;88(1):120-30. doi: 10.1128/JVI.02983-13. Epub 2013 Oct 16. J Virol. 2014. PMID: 24131715 Free PMC article.
References
-
- Ambroziak J A, Blackbourn D J, Herndier B G, Glogau R G, Gullett J H, McDonald A R, Lennette E T, Levy J A. Herpes-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science. 1995;268:582–583. - PubMed
-
- Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. - PubMed
-
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A, Gerhengorn M C. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

