Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides
- PMID: 10074111
- PMCID: PMC104021
- DOI: 10.1128/JVI.73.4.2658-2666.1999
Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides
Abstract
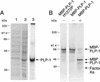
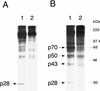
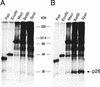
Proteolytic processing of the replicase gene product of mouse hepatitis virus (MHV) is essential for viral replication. In MHV strain A59 (MHV-A59), the replicase gene encodes two predicted papain-like proteinase (PLP) domains, PLP-1 and PLP-2. Previous work using viral polypeptide substrates synthesized by in vitro transcription and translation from the replicase gene demonstrated both cis and trans cleavage activities for PLP-1. We have cloned and overexpressed the PLP-1 domain in Escherichia coli by using a T7 RNA polymerase promoter system or as a maltose-binding protein (MBP) fusion protein. With both overexpression systems, the recombinant PLP-1 exhibited trans cleavage activity when incubated with in vitro-synthesized viral polypeptide substrates. Subsequent characterization of the recombinant PLP-1 revealed that in vitro trans cleavage is more efficient at 22 degrees C than at higher temperatures. Using substrates of increasing lengths, we observed efficient cleavage by PLP-1 requires a substrate greater than 69 kDa. In addition, when PLP-1 was expressed as a polypeptide that included additional viral sequences at the carboxyl terminus of the predicted PLP-1 domain, a fivefold increase in proteolytic activity was observed. The data presented here support previous data suggesting that in vitro and in vivo cleavage of the ORF 1a polyprotein by PLP-1 can occur in both in cis and in trans. In contrast to the cleavage activity demonstrated for PLP-1, no in vitro cleavage in cis or in trans could be detected with PLP-2 expressed either as a polypeptide, including flanking viral sequences, or as an MBP fusion enzyme.
Figures

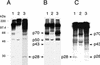

Similar articles
-
Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site.Virology. 1995 Jun 1;209(2):489-97. doi: 10.1006/viro.1995.1281. Virology. 1995. PMID: 7539970
-
Further in vitro characterization of mouse hepatitis virus papain-like proteinase 1: cleavage sequence requirements within pp1a.J Neurovirol. 2002 Apr;8(2):143-9. doi: 10.1080/13550280290049598. J Neurovirol. 2002. PMID: 11935466 Free PMC article.
-
Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59.J Virol. 1997 Feb;71(2):900-9. doi: 10.1128/JVI.71.2.900-909.1997. J Virol. 1997. PMID: 8995606 Free PMC article.
-
Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity.J Virol. 2004 Dec;78(24):13600-12. doi: 10.1128/JVI.78.24.13600-13612.2004. J Virol. 2004. PMID: 15564471 Free PMC article.
-
Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59.J Virol. 1995 Jun;69(6):3554-9. doi: 10.1128/JVI.69.6.3554-3559.1995. J Virol. 1995. PMID: 7745703 Free PMC article.
Cited by
-
The rubella virus nonstructural protease recognizes itself via an internal sequence present upstream of the cleavage site for trans-activity.Arch Virol. 2006 Sep;151(9):1841-51. doi: 10.1007/s00705-006-0744-9. Epub 2006 Mar 27. Arch Virol. 2006. PMID: 16570206 Free PMC article.
-
The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme.J Virol. 2005 Dec;79(24):15199-208. doi: 10.1128/JVI.79.24.15199-15208.2005. J Virol. 2005. PMID: 16306591 Free PMC article.
-
Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products.J Virol. 2000 Feb;74(4):1674-85. doi: 10.1128/jvi.74.4.1674-1685.2000. J Virol. 2000. PMID: 10644337 Free PMC article.
-
Functional analyses of the three simian hemorrhagic fever virus nonstructural protein 1 papain-like proteases.J Virol. 2014 Aug;88(16):9129-40. doi: 10.1128/JVI.01020-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899184 Free PMC article.
-
The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond.J Biol Chem. 2001 Aug 31;276(35):33220-32. doi: 10.1074/jbc.M104097200. Epub 2001 Jun 28. J Biol Chem. 2001. PMID: 11431476 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. pp. 10.1.1–10.1.3.
-
- Bonilla P J, Hughes S A, Piñón J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

