Osmosensing by bacteria: signals and membrane-based sensors
- PMID: 10066837
- PMCID: PMC98963
- DOI: 10.1128/MMBR.63.1.230-262.1999
Osmosensing by bacteria: signals and membrane-based sensors
Abstract
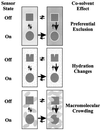
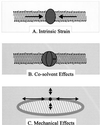
Bacteria can survive dramatic osmotic shifts. Osmoregulatory responses mitigate the passive adjustments in cell structure and the growth inhibition that may ensue. The levels of certain cytoplasmic solutes rise and fall in response to increases and decreases, respectively, in extracellular osmolality. Certain organic compounds are favored over ions as osmoregulatory solutes, although K+ fluxes are intrinsic to the osmoregulatory response for at least some organisms. Osmosensors must undergo transitions between "off" and "on" conformations in response to changes in extracellular water activity (direct osmosensing) or resulting changes in cell structure (indirect osmosensing). Those located in the cytoplasmic membranes and nucleoids of bacteria are positioned for indirect osmosensing. Cytoplasmic membrane-based osmosensors may detect changes in the periplasmic and/or cytoplasmic solvent by experiencing changes in preferential interactions with particular solvent constituents, cosolvent-induced hydration changes, and/or macromolecular crowding. Alternatively, the membrane may act as an antenna and osmosensors may detect changes in membrane structure. Cosolvents may modulate intrinsic biomembrane strain and/or topologically closed membrane systems may experience changes in mechanical strain in response to imposed osmotic shifts. The osmosensory mechanisms controlling membrane-based K+ transporters, transcriptional regulators, osmoprotectant transporters, and mechanosensitive channels intrinsic to the cytoplasmic membrane of Escherichia coli are under intensive investigation. The osmoprotectant transporter ProP and channel MscL act as osmosensors after purification and reconstitution in proteoliposomes. Evidence that sensor kinase KdpD receives multiple sensory inputs is consistent with the effects of K+ fluxes on nucleoid structure, cellular energetics, cytoplasmic ionic strength, and ion composition as well as on cytoplasmic osmolality. Thus, osmoregulatory responses accommodate and exploit the effects of individual cosolvents on cell structure and function as well as the collective contribution of cosolvents to intracellular osmolality.
Figures


Similar articles
-
Bacterial osmosensing: roles of membrane structure and electrostatics in lipid-protein and protein-protein interactions.Biochim Biophys Acta. 2004 Nov 3;1666(1-2):88-104. doi: 10.1016/j.bbamem.2004.06.013. Biochim Biophys Acta. 2004. PMID: 15519310 Review.
-
Bacterial osmosensing transporters.Methods Enzymol. 2007;428:77-107. doi: 10.1016/S0076-6879(07)28005-X. Methods Enzymol. 2007. PMID: 17875413 Review.
-
Osmosensing by bacteria.Sci STKE. 2006 Oct 17;2006(357):pe43. doi: 10.1126/stke.3572006pe43. Sci STKE. 2006. PMID: 17047223 Review.
-
Use of liposomes to study cellular osmosensors.Methods Mol Biol. 2010;606:21-30. doi: 10.1007/978-1-60761-447-0_3. Methods Mol Biol. 2010. PMID: 20013387
-
Osmosensing and osmoregulatory compatible solute accumulation by bacteria.Comp Biochem Physiol A Mol Integr Physiol. 2001 Oct;130(3):437-60. doi: 10.1016/s1095-6433(01)00442-1. Comp Biochem Physiol A Mol Integr Physiol. 2001. PMID: 11913457 Review.
Cited by
-
Osmotic Stress Confers Enhanced Cell Integrity to Hydrostatic Pressure but Impairs Growth in Alcanivorax borkumensis SK2.Front Microbiol. 2016 May 18;7:729. doi: 10.3389/fmicb.2016.00729. eCollection 2016. Front Microbiol. 2016. PMID: 27242746 Free PMC article.
-
Osmotic stress signaling and osmoadaptation in yeasts.Microbiol Mol Biol Rev. 2002 Jun;66(2):300-72. doi: 10.1128/MMBR.66.2.300-372.2002. Microbiol Mol Biol Rev. 2002. PMID: 12040128 Free PMC article. Review.
-
High Osmolarity Modulates Bacterial Cell Size through Reducing Initiation Volume in Escherichia coli.mSphere. 2018 Oct 24;3(5):e00430-18. doi: 10.1128/mSphere.00430-18. mSphere. 2018. PMID: 30355666 Free PMC article.
-
Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate.Biochemistry. 2010 May 25;49(20):4361-73. doi: 10.1021/bi100092a. Biochemistry. 2010. PMID: 20201585 Free PMC article.
-
Osmolality, temperature, and membrane lipid composition modulate the activity of betaine transporter BetP in Corynebacterium glutamicum.J Bacteriol. 2007 Oct;189(20):7485-96. doi: 10.1128/JB.00986-07. Epub 2007 Aug 10. J Bacteriol. 2007. PMID: 17693504 Free PMC article.
References
-
- Alemohammad M M, Knowles C J. Osmotically induced volume and turbidity changes of Escherichia coli due to salts, sucrose and glycerol, with particular reference to the rapid permeation of glycerol into the cell. J Gen Microbiol. 1974;82:125–142. - PubMed
-
- Alexander, D., A. Lipski, and J. M. Wood. Unpublished data.
-
- Altendorf K, Epstein W. Kdp-ATPase of Escherichia coli. Cell Physiol Biochem. 1993;4:160–168.
-
- Anderson C F, Record M T., Jr Salt-nucleic acid interactions. Annu Rev Phys Chem. 1995;46:657–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

