Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences
- PMID: 10022858
- PMCID: PMC83964
- DOI: 10.1128/MCB.19.3.1705
Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences
Abstract
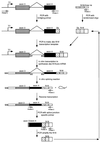
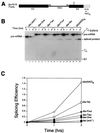
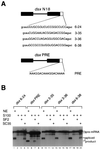
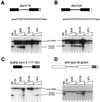
Splicing enhancers are RNA sequences required for accurate splice site recognition and the control of alternative splicing. In this study, we used an in vitro selection procedure to identify and characterize novel RNA sequences capable of functioning as pre-mRNA splicing enhancers. Randomized 18-nucleotide RNA sequences were inserted downstream from a Drosophila doublesex pre-mRNA enhancer-dependent splicing substrate. Functional splicing enhancers were then selected by multiple rounds of in vitro splicing in nuclear extracts, reverse transcription, and selective PCR amplification of the spliced products. Characterization of the selected splicing enhancers revealed a highly heterogeneous population of sequences, but we identified six classes of recurring degenerate sequence motifs five to seven nucleotides in length including novel splicing enhancer sequence motifs. Analysis of selected splicing enhancer elements and other enhancers in S100 complementation assays led to the identification of individual enhancers capable of being activated by specific serine/arginine (SR)-rich splicing factors (SC35, 9G8, and SF2/ASF). In addition, a potent splicing enhancer sequence isolated in the selection specifically binds a 20-kDa SR protein. This enhancer sequence has a high level of sequence homology with a recently identified RNA-protein adduct that can be immunoprecipitated with an SRp20-specific antibody. We conclude that distinct classes of selected enhancers are activated by specific SR proteins, but there is considerable sequence degeneracy within each class. The results presented here, in conjunction with previous studies, reveal a remarkably broad spectrum of RNA sequences capable of binding specific SR proteins and/or functioning as SR-specific splicing enhancers.
Figures


Similar articles
-
The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20.J Biol Chem. 2003 Aug 29;278(35):32943-53. doi: 10.1074/jbc.M301090200. Epub 2003 Jun 24. J Biol Chem. 2003. PMID: 12826680
-
Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA.Mol Cell Biol. 1999 Jan;19(1):261-73. doi: 10.1128/MCB.19.1.261. Mol Cell Biol. 1999. PMID: 9858550 Free PMC article.
-
The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers.RNA. 1999 Mar;5(3):468-83. doi: 10.1017/s1355838299981967. RNA. 1999. PMID: 10094314 Free PMC article.
-
SR proteins as potential targets for therapy.Prog Mol Subcell Biol. 2006;44:65-87. doi: 10.1007/978-3-540-34449-0_4. Prog Mol Subcell Biol. 2006. PMID: 17076265 Review.
-
Plant serine/arginine-rich proteins: versatile players in RNA processing.Planta. 2023 May 5;257(6):109. doi: 10.1007/s00425-023-04132-0. Planta. 2023. PMID: 37145304 Review.
Cited by
-
Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.EMBO J. 2006 Nov 1;25(21):5126-37. doi: 10.1038/sj.emboj.7601385. Epub 2006 Oct 12. EMBO J. 2006. PMID: 17036044 Free PMC article.
-
Single nucleotide polymorphism-based validation of exonic splicing enhancers.PLoS Biol. 2004 Sep;2(9):E268. doi: 10.1371/journal.pbio.0020268. Epub 2004 Aug 31. PLoS Biol. 2004. PMID: 15340491 Free PMC article.
-
SRSF7 and SRSF3 depend on RNA sequencing motifs and secondary structures to regulate Microprocessor.Life Sci Alliance. 2023 Feb 7;6(4):e202201779. doi: 10.26508/lsa.202201779. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36750366 Free PMC article.
-
The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element.EMBO J. 2001 Oct 15;20(20):5748-58. doi: 10.1093/emboj/20.20.5748. EMBO J. 2001. PMID: 11598017 Free PMC article.
-
Sequence information for the splicing of human pre-mRNA identified by support vector machine classification.Genome Res. 2003 Dec;13(12):2637-50. doi: 10.1101/gr.1679003. Genome Res. 2003. PMID: 14656968 Free PMC article.
References
-
- Abmayr S M, Workman J L. Preparation of nuclear and cytoplasmic extracts from mammalian cells. In: Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. pp. 12.1.1–12.1.9.
-
- Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. - PubMed
-
- Amrein H, Gorman M, Nöthiger R. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. - PubMed
-
- Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer-2. Cell. 1994;76:735–746. - PubMed
-
- Baker B S. Sex in flies: the splice of life. Nature. 1989;340:521–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
