Abstract
Tie2 is an endothelium-specific receptor tyrosine kinase that is required for both normal embryonic vascular development and tumor angiogenesis and is thought to play a role in vascular maintenance. However, the signaling pathways responsible for the function of Tie2 remain unknown. In this report, we demonstrate that the p85 subunit of phosphatidylinositol 3-kinase (PI3-kinase) associates with Tie2 and that this association confers functional lipid kinase activity. Mutation of tyrosine 1101 of Tie2 abrogated p85 association both in vitro and in vivo in yeast. Tie2 was found to activate PI3-kinase in vivo as demonstrated by direct measurement of increases in cellular phosphatidylinositol 3-phosphate and phosphatidylinositol 3,4-bisphosphate, by plasma membrane translocation of a green fluorescent protein-Akt pleckstrin homology domain fusion protein, and by downstream activation of the Akt kinase. Activation of PI3-kinase was abrogated in these assays by mutation of Y1101 to phenylalanine, consistent with a requirement for this residue for p85 association with Tie2. These results suggest that activation of PI3-kinase and Akt may in part account for Tie2’s role in both embryonic vascular development and pathologic angiogenesis, and they are consistent with a role for Tie2 in endothelial cell survival.
Tie2 (also called Tek) is a member of a novel family of receptor tyrosine kinases (RTKs) (16, 17, 37, 42, 72) that are expressed predominantly on endothelial cells or their embryonic precursors (14, 16, 17, 37, 42) and that are required for normal vascular development (15, 52, 55). Functional disruption of Tie2 in transgenic mice results in embryonic lethality by day E9.5 to 10.5 with effects on the microvasculature resulting in reduced numbers of endothelial cells and abnormalities of vascular morphogenesis (15, 55). Knockout of the activating Tie2 ligand, angiopoietin-1 (Ang1), or overexpression of a related, inhibitory ligand, angiopoietin-2 (Ang2), resulted in phenotypes similar to the Tie2 knockout (43, 64). Taken together, these findings suggest a role for Tie2 in endothelial cell maintenance, survival, and/or vascular morphogenesis (24).
In addition to a role in embryonic vascular development, data from our laboratory suggest that Tie2 plays an important role in the adult vasculature. For example, Tie2 expression is increased in the vasculature of malignant breast tumors (49), and a soluble extracellular domain of Tie2 inhibits tumor angiogenesis and growth (39). Tie2 is also broadly expressed and tyrosine phosphorylated in a variety of adult tissues in which the endothelium is normally quiescent (69). These findings are consistent with a dual role for Tie2 in both the growth and the maintenance of the adult vasculature.
To better understand the role of Tie2 in vascular growth and maintenance, it will be important to identify the signal transduction pathways responsible for these functions. Currently, little is known about the specific signaling proteins and pathways utilized by Tie2. We demonstrated previously that Tie2 associates with the Src homology 2 (SH2) domains of the adapter protein Grb2 and the protein tyrosine phosphatase Shp2/SH-PTP2 (31). Although both of these proteins have been linked to activation of Ras and mitogen-activated protein (MAP) kinase (20, 56, 66), Tie2 does not appear to activate MAP kinase (36) or stimulate cellular proliferation (10, 36). To identify other proteins and signaling pathways downstream of Tie2, the Tie2 kinase domain was used as a bait to screen a human fetal heart cDNA library with the yeast two-hybrid system. Here we report the association of the p85 regulatory subunit of phosphatidylinositol 3-kinase (PI3-kinase) with a nonconsensus binding motif on Tie2 and demonstrate, by both a novel approach and high-pressure liquid chromatography (HPLC) analysis of phospholipids, that stimulation of Tie2 activates PI3-kinase in vivo. Furthermore, stimulation of Tie2 results in activation of Akt/protein kinase B, a process that has been linked to cell survival and antiapoptosis (9, 11, 38) and that may in part account for Tie2’s role in vascular growth and maintenance.
(This work was presented in part at the 69th Annual Scientific Sessions of the American Heart Association, New Orleans, La., November 1996 [34a].)
MATERIALS AND METHODS
Human fetal heart cDNA library construction.
cDNA synthesis was performed with the Stratagene cDNA synthesis kit according to the manufacturer’s instructions with a slight modification for library construction. Briefly, poly(A)+ mRNA derived from human fetal heart (Clontech) was used to direct the synthesis of first-strand cDNA by Moloney murine leukemia virus reverse transcriptase with an oligo(dT)-XhoI linker-primer. Following second-strand synthesis, the cDNA was ligated to an EcoRI adapter, digested with XhoI, and subsequently size selected. Fractions containing cDNAs ranging from 500 bp to >6 kb in length were pooled, directionally subcloned into the yeast library plasmid pJG4-5 (kindly provided by Roger Brent, Harvard University, Cambridge, Mass.) (22, 23), and introduced into electrocompetent DH10B Escherichia coli by electroporation. Evaluation of the number of recombinant clones and of the size range of the cDNA inserts was accomplished by restriction enzyme digestion analysis of plasmids isolated from 20 randomly picked bacterial transformants. Analysis demonstrated that 90% (18 of 20) of these plasmids contained cDNAs with insert sizes ranging from 0.7 to 3.5 kb (average, ∼1.3 kb). The resultant plasmid cDNA library contains 9 × 106 primary recombinant clones encoded as fusion proteins with the B42 transcriptional activation domain under the direction of the GAL1 promoter.
Generation and testing of bait plasmids and yeast two-hybrid screening.
All yeast plasmid vectors and yeast strains were generously provided by Roger Brent. Cloning of the murine Tie2 kinase domain and site-directed mutagenesis to generate a kinase-inactive mutant (K854R) and specific tyrosine-to-phenylalanine mutations (Y1067F, Y1101F, Y1112F, and the double mutant Y2F, consisting of both Y1101F and Y1112F) have been described previously (31). The entire murine Tie1 kinase domain cDNA was generated by PCR from Py-4-1 endothelial cell cDNA (12). These cDNAs were then subcloned into the vector pJK202 (22) to generate the bait plasmids lexA-Tie1 and lexA-Tie2 (with or without specific mutations). Baits were tested for inappropriate transcriptional activation of the reporter genes LEU2 and lacZ in the yeast strains EGY48/ pSH18-34 and EGY191/pJK103 as described elsewhere (19, 22). All yeast transformations were performed with the Yeastmaker yeast transformation kit (Clontech) according to the manufacturer’s instructions, and transformants were selected by growth on appropriate yeast dropout media (Bio 101). Because lexA-Tie2 weakly activated transcription of LEU2 in EGY48/pSH18-34, library screening was performed in the strain EGY191/pJK103, which contains fewer lexA operator elements upstream of its reporter genes (19). Interactions with lexA-Tie1 were tested in the EGY48/pSH18-34 strain. Yeast cells expressing lexA-Tie2 were transformed with 1 μg of human fetal heart cDNA, and two-hybrid screening was performed as previously described (22, 23). Yeast colonies that demonstrated galactose-dependent activation of both reporters (blue staining in the presence of X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] and leucine-independent growth) were considered true positive interactors and were further evaluated.
Categorization and specificity testing of interactors.
Library plasmids from yeast colonies expressing Tie2-interacting proteins were rescued by transformation of yeast plasmid DNA into KC8 E. coli followed by selection on minimal medium lacking tryptophan (22). Simultaneously, to simplify analysis and categorization of multiple interacting plasmids, interactor cDNAs were isolated by PCR with the following oligonucleotide primers specific for flanking sequences within pJG4-5: 5′ oligonucleotide, 5′-CCAGCCTCTTGCTGAGTGGAG-3′; 3′ oligonucleotide, 5′-GACAAGCCGACAACCTTGATTGT-3′. PCR-generated interactor cDNA fragments were agarose gel purified and categorized by restriction enzyme digestion with HinfI and HhaI. Specificity of the interaction of library-expressed proteins with Tie2 was tested by retransforming 100 ng of interactor plasmid into yeast expressing lexA-Tie2 and into strains expressing the unrelated bait plasmid pRFHM1 (encoding a portion of the bicoid protein) (22), pKREV (the small G protein Rap1A), or pSCRPB (subunit RPB7 of Saccharomyces cerevisiae RNA polymerase II) (19) and the related bait plasmids lexA-Tie1 and lexA-Tie2-K854R. Transformants were selected by plating onto Ura− His− Trp− medium containing glucose, and six colonies from each transformation were picked and replica plated as described above for two-hybrid screening. Interaction was judged specific if galactose-dependent transcriptional activation of LEU2 and lacZ was demonstrated in strains expressing only related baits and not in strains expressing any of the three unrelated baits. cDNA from specific interactors was sequenced and evaluated by the Basic Local Alignment Search Tool through the National Center for Biotechnology Information Internet site (www.ncbi.nlm.nih.gov).
Generation of recombinant GST-kinase fusion proteins.
Recombinant Tie2 kinases, with or without specific mutations, were expressed as fusion proteins with glutathione S-transferase (GST) in Sf9 insect cells as previously described (31).
In vitro association assays.
Association assays were performed essentially as described elsewhere (31) with slight modifications. Approximately 500 ng of each recombinant kinase was purified on glutathione-Sepharose (Pharmacia) in 500 μl of lysis buffer (137 mM NaCl, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 20 mM Tris-HCl, pH 8.0). Glutathione beads and immobilized kinases were washed three times with ice-cold lysis buffer and then subjected to an in vitro kinase reaction by incubation at room temperature for 20 min in 100 μl of kinase buffer (100 mM NaCl, 12 mM MgCl2, 1 mM dithiothreitol, 20 mM Tris-HCl, pH 7.5) containing 1 mM ATP. The autophosphorylated kinases were incubated at 4°C for 2 h with 1 ml of endothelial cell lysate, and the beads and associated proteins were washed and eluted by boiling into Laemmli sample buffer. Associated proteins were separated by sodium dodecyl sulfate (SDS)–8 to 16% polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and blotted with anti-Tie2 polyclonal antibody (PAb) (1:10,000) (31), antiphosphotyrosine monoclonal antibody (MAb) (1:1,000, clone PY20; Transduction Laboratories), anti-p85 MAb (1:5, clone N7B; generously provided by Anke Klippel, Chiron Corp., Emeryville, Calif.), or anti-Grb2 MAb (1:5,000, clone 81; Transduction Laboratories). To generate endothelial cell lysates, EAhy.926 cells, an endothelial fusion cell line (18), were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml (all Gibco BRL) and HAT supplement (hypoxanthine, aminopterin, and thymidine [Sigma]) at 37°C in a 5% CO2 atmosphere. Confluent cells in 100-mm dishes were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in 1 ml of cold lysis buffer containing protease inhibitors (1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate [vanadate]).
Peptide competition assays.
Peptides were generated as described previously (31). Peptides were 14-mers derived from sequences flanking tyrosines 1101 and 1112 in the carboxy terminus of Tie2. The sequence of peptide pY1101 was EERKTpYVNTTLYEK, and that of peptide pY1112 was YEKFTpYAGIDCSAE. The corresponding unphosphorylated peptides were used as controls. For peptide competition experiments, in vitro association assays were performed with recombinant GST-Tie2 and EAhy.926 lysates as described above except that endothelial cell lysates were preincubated for 30 min at 4°C with increasing concentrations of peptide prior to the addition of cell lysates to autophosphorylated GST-Tie2 immobilized on glutathione-Sepharose. Tie2-associated proteins were evaluated by SDS-PAGE and Western blotting as described for association assays.
PI3-kinase assays.
To assay Tie2 kinase-associated PI3-kinase activity, in vitro association assays were performed with recombinant GST-kinases and endothelial cell lysates as described above, but after incubation and association at 4°C, the glutathione beads and associated proteins were washed twice with ice-cold lysis buffer and twice with 20 mM Tris-HCl, pH 7.4. After the final wash, the beads and associated proteins were incubated on ice for 10 min with 10 μg of sonicated PI (Sigma) in 20 mM Tris-HCl, pH 7.4. Kinase reactions were initiated by the addition of 40 μl of kinase buffer (30 μM MgCl2, 10 μM unlabeled ATP, 20 mM Tris-HCl, pH 7.4) containing 10 μCi of [γ-32P]ATP (6,000 Ci/mmol; Amersham) and 20 μM adenosine (to inhibit PI4-kinase activity) and were incubated at room temperature for 20 min (34, 68). Reactions were stopped by the addition of 1 M HCl, and then lipids were extracted into a 1:1 mixture of chloroform and methanol and spotted onto Silica gel 60 plates (Merck) pretreated with 1% potassium oxalate in methanol (3:2) and separated by one-dimensional thin-layer chromatography in a developing solution of chloroform, methanol, water, and ammonium hydroxide (18:14:3:1). Conversion of PI to PI3-phosphate (PI3P) was visualized by autoradiography and quantitated with a Molecular Dynamics PhosphorImager screen and ImageQuant software, version 4.00 (Molecular Dynamics). Baseline activity in each experiment was assessed from incubation of endothelial cell lysates with glutathione-Sepharose alone (without recombinant kinase). Results in each experiment were corrected for this baseline activity and expressed relative to wild-type Tie2 kinase-associated PI3-kinase activity.
Cell lines and transfections.
NIH 3T3 cells were obtained from the American Type Culture Collection. GP+E86 retroviral packaging cells were a gift from Arthur Bank, Columbia University, New York, N.Y. (44). Both cell lines were maintained in DMEM supplemented with 10% FBS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 37°C in a 5% CO2 atmosphere. A chimeric receptor consisting of the extracellular and transmembrane domains of the colony-stimulating factor 1 (CSF-1) receptor, c-fms (provided by Charles Sherr, St. Jude’s Children’s Hospital, Memphis, Tenn.), and the cytoplasmic kinase domain of Tie2 (designated fTie2) was generated in the mammalian retroviral expression vector LNCX6 (45). The previously described tyrosine-to-phenylalanine mutants and the kinase-inactive mutant of Tie2 (31) were engineered into fTie2 in LNCX6. The resultant plasmids were transfected into GP+E86 packaging cells with Lipofectamine (Gibco BRL) according to the manufacturer’s instructions. Stably transfected cell lines were selected with 400 μg of G418 (Geneticin; Gibco BRL) per ml, and clonal populations were screened for fTie2 expression by Western blotting of cell lysates with a Tie2-specific PAb (31). From these stable cell lines, tissue culture supernatants (1 ml) containing recombinant ecotropic retroviruses were used to infect NIH 3T3 fibroblasts in 4 ml of Opti-MEM I (Gibco BRL) containing 8 μg of hexadimethrine bromide (Polybrene; Sigma) per ml for 4 to 6 h at 37°C. Stably transfected 3T3 cells were selected and analyzed for fTie2 expression as described above and subsequently maintained in the presence of 400 μg of G418 per ml.
Immunoprecipitation and Western blotting.
Untransfected NIH 3T3 cells or cells stably expressing fTie2 chimeric receptors, with or without specific point mutations, were grown in 100-mm dishes to subconfluence and then serum starved overnight in DMEM–0.2% FBS. Cells were washed once with PBS and then stimulated for 10 min at 37°C with 500 ng of CSF-1 (Genetics Institute, Cambridge, Mass.) per ml or 30 ng of platelet-derived growth factor BB (PDGF-BB) (R&D Systems) per ml in Dulbecco’s PBS (Gibco BRL) containing 1 mM vanadate. Cells were washed twice with PBS containing 0.5 mM vanadate and then lysed as described for EA.hy926 cells. Chimeric receptor proteins were immunoprecipitated from cell lysates with a rat MAb against c-fms (1:200) (clone 3-4A4-E4; Oncogene Science) and immobilized on protein G agarose (Santa Cruz Biotechnology). Protein complexes were washed with lysis buffer, eluted by boiling into Laemmli sample buffer, and separated by SDS-PAGE. After transfer to nitrocellulose, proteins were analyzed by Western blotting with anti-Tie2 PAb (1:10,000), antiphosphotyrosine (1:1,000), or a goat PAb specific for the C terminus of Akt1 (1:1,000) (C-20; Santa Cruz Biotechnology).
Generation and analysis of GFP-Akt PH domain fusion proteins in vivo.
The pleckstrin homology (PH) domain of human c-Akt (amino acids 1 to 147) was subcloned in-frame and 3′ of cycle 3 green fluorescent protein (GFP) (6) in the eukaryotic RNA expression vector Hiro3 (70). An additional S65T mutation was added to cycle 3 GFP to increase its fluorescence intensity (26). GFP-PH RNA was sequentially transcribed in vitro and polyadenylated (70) and then electroporated into either untransfected NIH 3T3 cells or those expressing fTie2 chimeric receptors, as described previously (59, 60, 65). Electroporated cells were allowed to express the GFP-tagged PH domains for 3 h, during which time they were briefly serum starved in DMEM and then visualized by confocal laser scanning microscopy (Zeiss LSM) before and after stimulation with CSF-1 (500 ng/ml; Genetics Institute) or PDGF-BB (40 ng/ml; R&D Systems), both in the presence of 1 mM vanadate. As a control, cells were also stimulated with vanadate alone. Each cell line was also evaluated as described above following pretreatment with 50 nM wortmannin (Sigma) to inhibit PI3-kinase activity. The relative change in fluorescence at the plasma membrane compared to that of the cytosol was analyzed with a fluorescence intensity plot of cross sections through cells in confocal midsections. The ratio was determined according to the formula (p − c)/(c + p), where p is the fluorescence intensity at the plasma membrane and c is the fluorescence intensity in the cytosol (59). Analysis was performed on 10 to 20 cells in each experiment, and results were expressed as the means ± standard errors of the means (SEM), as previously described (59).
Akt kinase assays.
To assay Akt kinase activity, cell lysates prepared as described above were incubated overnight with anti-Akt PAb (1:200), and immune complexes were immobilized on protein A agarose (Santa Cruz Biotechnology) for 1 h at 4°C. The beads and protein complexes were washed as described by Klippel et al. (34) and then incubated for 20 min at room temperature in 30 μl of kinase buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 10 mM MnCl2, 1 mM dithiothreitol, 10 mM β-glycerophosphate, 1 μM protein kinase A inhibitor peptide [Santa Cruz Biotechnology], 50 μM unlabeled ATP) containing 5 μCi of [γ-32P]ATP (6,000 Ci/mmol; Amersham) and 1.5 μg of histone H2B (5 mg/ml in water; Boehringer Mannheim) per sample. Reactions were stopped by the addition of 10 μl of 4× Laemmli sample buffer with 5% β-mercaptoethanol, and samples were boiled for 5 min. Proteins were separated by SDS–15% PAGE, and incorporation of 32P into histone H2B was evaluated by autoradiography.
Measurement of PI phosphates in vivo.
Cells expressing either wild-type or mutant fTie2 receptors were plated in 10-cm dishes and labeled with 5 μCi of [3H]myo-inositol (20 Ci/mmol; American Radiolabeled Chemicals, Inc., St. Louis, Mo.) per ml for 72 h, as described elsewhere (71). The cells were then starved for 2 h in serum-free DMEM, washed once with PBS, and then either left unstimulated or stimulated with CSF-1 for 8 min in the presence of 1 mM sodium orthovanadate, as described above. After stimulation, the media were aspirated and cells were scraped into 1 ml of 1 M HCl. The cellular debris was pelleted by centrifugation, and lipids were extracted by resuspension in 100 μl of 0.5 M HCl and 372 μl of CHCl3-methanol (1:2 [vol/vol]) by vigorous reciprocal shaking in a Mini Bead-Beater (BioSpec, Inc., Bartlesville, Okla.) with 100 μl of acid-washed glass beads (425- to 600-μm diameter; Sigma). After the addition of 125 μl of CHCl3 and 125 μl of 2 M KCl, the samples were vortexed for 1 min and then centrifuged for 5 min at 20,800 × g. The lipid-containing organic phase was transferred to a screw-cap polypropylene tube along with 50 μl of brain extract (Sigma) and dried under vacuum. The PI phosphates were then deacylated, and the resultant glycerophosphatidylinositol (gPI) phosphates were extracted into aqueous solution as described by Stolz et al. (63) and stored at −80°C until HPLC analysis. Samples were equilibrated to 10 mM ammonium phosphate (AP), pH 3.5; applied to a Partisphere SAX 10-μm column (4.6 × 250 mm; Whatman); and eluted along a linear gradient of 10 to 340 mM AP over 30 min, 340 mM to 1.02 M AP over 15 min, 1.02 to 1.7 M AP over 5 min, and constant 1.7 M AP for 10 min. The radioactivity of the eluate was measured with a BetaRAM in-line detector (INUS, Tampa, Fla.) and analyzed with the manufacturer’s software. 32P-labeled PI(3,4)P2 and PI(3,4,5)P3 standards (kindly provided by Phil Majerus, Washington University, St. Louis, Mo.) were deacylated as described elsewhere (63) and resolved and analyzed as described above. Samples were run in triplicate, and counts per minute corresponding to each gPI phosphate (gPIP) peak were quantitated. For each sample, background counts per minute were estimated from the baseline on the chromatogram and subtracted from the total counts per minute for each peak. The resultant total counts per minute corresponding to each phospholipid peak was divided by the total cellular gPI counts per minute in each sample, and means ± SEM of three separate experiments were expressed relative to total cellular gPI. Statistical analysis was performed with the paired two-sample Student t test.
RESULTS
The C-terminal SH2 domain of p85 associates with Tie2 in yeast.
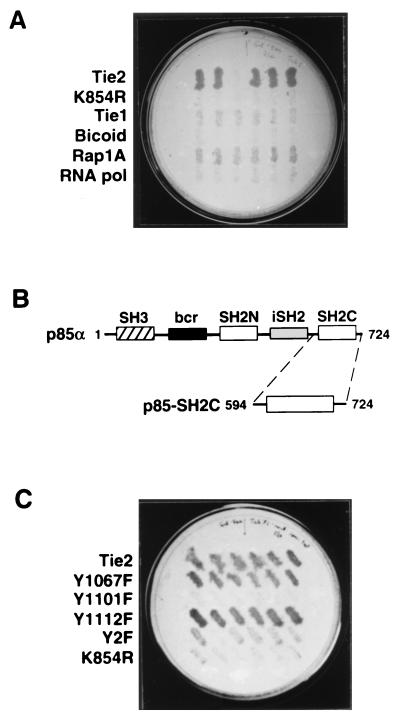
Because of the apparent role of Tie2 in vascular and endocardial development (15, 55), we reasoned that embryonic tissues undergoing active angiogenesis would be rich in endothelial signaling proteins that interact with Tie2. Therefore, with the yeast two-hybrid system, we screened a human fetal heart cDNA library (22, 23) with a lexA-Tie2 kinase bait to identify potential downstream interacting proteins. From an initial screen of approximately 106 primary yeast transformants, 8 interactors were identified by simultaneous LEU2 and lacZ transcription. Six of these eight interactors contained 1.3-kb cDNA inserts with identical restriction digestion patterns (data not shown). This cDNA encoded a protein that interacted strongly with the Tie2 kinase in yeast, as demonstrated by leucine-independent growth (Fig. 1A). Notably, this interaction was dependent on Tie2 autophosphorylation because no interaction was demonstrated with a kinase-deficient mutant of Tie2 (K854R). In addition, the interaction was specific for Tie2, since no appreciable interaction was detected with the closely related Tie1 kinase or with any of three unrelated bait proteins (Fig. 1A). Similar results were observed for lacZ transcription (data not shown).
FIG. 1.
Identification of p85 as a specific Tie2 interactor by the yeast two-hybrid system. (A) Specific, autophosphorylation-dependent interaction of a human fetal heart library-encoded protein with Tie2. A plasmid encoding a 1.3-kb Tie2 interactor, from a fetal heart cDNA library, was transformed into yeast expressing the original lexA-Tie2 kinase bait, lexA-Tie2-K854R, lexA-Tie1 kinase, or one of three unrelated lexA bait proteins (bicoid protein, the small G protein Rap1A, and subunit RPB7 of S. cerevisiae RNA polymerase II). Six colonies from each transformation (horizontal rows of colonies) were picked and replica plated as described in Materials and Methods and tested for galactose-dependent activation of LEU2 on medium lacking leucine. (B) Schematic representation of structural domains within the full-length p85α and p85-SH2C. cDNA sequencing demonstrated that the 1.3-kb interactor cDNA encoded the entire C-terminal SH2 domain of p85α (p85-SH2C) (first and last amino acid residues of each protein are shown). bcr, breakpoint cluster region; iSH2, inter-SH2 domain. (C) Mutational analysis of the p85-SH2C–Tie2 interaction. The plasmid encoding p85-SH2C was transformed into yeast expressing either wild-type or mutant lexA-Tie2 kinase baits. Six colonies from each transformation were replica plated as described for panel A and tested for galactose-dependent LEU2 transcription. Y2F, Y1101F plus Y1112F.
Sequence analysis demonstrated that the 1.3-kb cDNA insert from this interactor plasmid encoded amino acids 594 to 724 of p85α (57), the regulatory subunit of PI3-kinase (Fig. 1B), with approximately 900 bp of 3′ untranslated sequence. This region of p85 encompasses the entire C-terminal SH2 domain (p85-SH2C) and a small region flanking the inter-SH2 domain (iSH2), which mediates interaction with p110 (30, 32), the catalytic subunit of PI3-kinase (27) (Fig. 1B). Interestingly, the interacting domain included a 9-amino-acid insert that replaces amino acid 604 of p85α. This insert has been shown to be the product of alternative splicing of p85α, and this variant is expressed predominantly in heart and skeletal muscle (2).
p85-SH2C associates with a nonconsensus binding motif at Y1101.
Interaction of SH2 domains with RTKs is known to be phosphotyrosine dependent (20, 56, 66). Having confirmed that the interaction of p85-SH2C with Tie2 was autophosphorylation dependent by using the K854R mutant bait, we performed additional mutational analysis to determine whether this interaction was mediated by a specific tyrosine residue on Tie2. Based on previous work, two tyrosine residues in the C tail of Tie2, Y1101 and Y1112, are thought to be important autophosphorylation sites (31). Although neither Y1101 nor Y1112 conforms to the known p85 consensus binding motif YXXM (58), Y1067 within the kinase domain contains the motif YDLM; thus, the Y1067F mutant bait was also evaluated for p85-SH2C interaction. Mutation of either Y1067 or Y1112, the Shp2 binding site (31), had no appreciable effect on Tie2–p85-SH2C interaction (Fig. 1C). In contrast, mutation of the nonconsensus Y1101 to phenylalanine, either alone or in combination with Y1112F (Y2F), appeared to almost completely abrogate interaction. Similar results were noted for lacZ transcription (data not shown).
Full-length p85 from endothelial cells preferentially associates with the Tie2 kinase in vitro at Y1101.
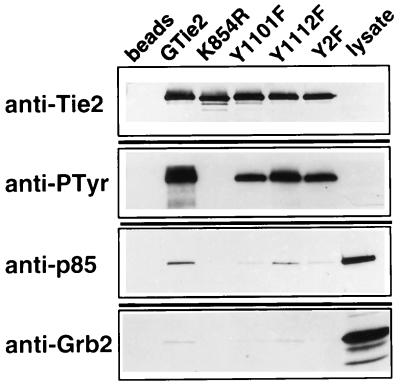
Using wild-type or mutant GST-Tie2 kinase fusion proteins in an in vitro association assay with endothelial cell lysates, we next evaluated whether p85 from endothelial cells could bind to the Tie2 kinase in vitro and whether the full-length protein, with two SH2 domains, would demonstrate a similar binding site preference for Y1101. Western blotting of recombinant kinases and associated proteins demonstrated a lack of phosphorylation of K854R, confirming that phosphorylation of Tie2 in vitro is an autophosphorylation event and is not mediated by another kinase from either endothelial cell or insect cell lysates (Fig. 2). Notably, the Y1112F mutant, which fails to bind the tyrosine phosphatase Shp2, demonstrates the greatest autophosphorylation of the mutants, as shown previously (31). Anti-p85 immunoblots demonstrated association of p85 with the wild-type Tie2 kinase but not with K854R (Fig. 2), confirming that the interaction is phosphotyrosine dependent. Similar to the results for yeast, the Y1101F mutation, either alone or in combination with Y1112F (Y2F), markedly diminished p85 association with Tie2. However, Tie2-p85 association was only mildly reduced by mutation of Y1112 alone (Fig. 2). These results demonstrate that full-length p85 associates with the autophosphorylated Tie2 kinase in vitro and that the major binding site is tyrosine 1101, as demonstrated in the two-hybrid system.
FIG. 2.
Endothelial p85 associates preferentially with Y1101 of the Tie2 kinase in vitro. Similar amounts of recombinant GST-Tie2 kinase fusion proteins, with or without specific mutations, were immobilized on glutathione-Sepharose, subjected to in vitro kinase reactions, and incubated with endothelial cell lysates. Kinase-associated proteins were separated by SDS–8 to 16% PAGE and analyzed by Western blotting with antibodies against Tie2, phosphotyrosine (PTyr), p85, and Grb2. An aliquot of crude endothelial cell lysate (lysate) was used as a comparison for p85 and Grb2, and glutathione-Sepharose alone, without recombinant kinase, was incubated with endothelial cell lysates as a negative control (beads).
Because Grb2 has been shown to associate with Y1101 of Tie2 (31), the blots of GST-Tie2-associated proteins were simultaneously probed with an antibody to Grb2. The similar patterns of binding to Tie2 by both p85 and Grb2 (Fig. 2) suggest that these two SH2 domain-containing proteins share the Y1101 binding site on Tie2, similar to findings with the PDGF β receptor and the hepatocyte growth factor receptor (47, 50).
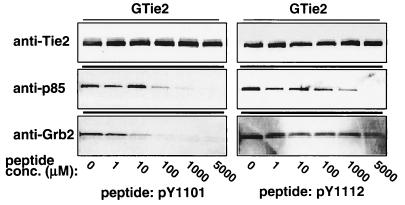
A phosphopeptide mimicking Y1101, but not Y1112, specifically inhibits p85-Tie2 association.
To demonstrate that the interaction of p85 with Tie2 was specific for Y1101 and that changes in association were not just due to decreases in receptor phosphorylation caused by mutation of individual tyrosine residues, synthetic peptides corresponding to sequences flanking tyrosines 1101 and 1112 were used in peptide competition experiments. Consistent with previous data (31), association of Grb2 with Tie2 was effectively inhibited with 1 to 10 μM phosphopeptide pY1101 (Fig. 3, left panels). Similarly, binding of p85 to GST-Tie2 was inhibited with between 10 and 100 μM pY1101 peptide (Fig. 3, left panels) but not with the unphosphorylated peptide (data not shown). In contrast, a phosphopeptide derived from sequences flanking tyrosine 1112 (pY1112) was approximately 50- to 100-fold less effective at competing for Tie2 binding (Fig. 3, right panels) while the corresponding unphosphorylated peptide (Y1112) did not affect p85-Tie2 association (data not shown). Neither the pY1112 nor the Y1112 peptide inhibited Grb2-Tie2 interaction at the concentrations tested (Fig. 3, right panels, and data not shown). These results confirm that preferential association of p85 with autophosphorylated Tie2 at Y1101 is specific for this phosphotyrosine residue, although they do not entirely rule out a secondary site of association at Y1112. These findings remain consistent with possible competition between p85 and Grb2 for binding to Tie2, and they suggest that p85 may bind Tie2 with slightly lower affinity than does Grb2.
FIG. 3.
Inhibition of p85-Tie2 kinase association with a synthetic phosphopeptide mimicking tyrosine 1101. In vitro association assays were performed as described for Fig. 2 except that endothelial cell lysates were preincubated with increasing concentrations of synthetic phosphopeptides derived from sequences flanking Y1101 (pY1101, left panels) or Y1112 (pY1112, right panels). GST-Tie2-associated p85 and Grb2 were separated by SDS–8 to 16% PAGE and analyzed by Western blotting as described for Fig. 2. Blots were probed with anti-Tie2 to demonstrate equal loading of GST-Tie2 in each lane (upper panels).
Association of p85 with Tie2 confers association of PI3-kinase activity.
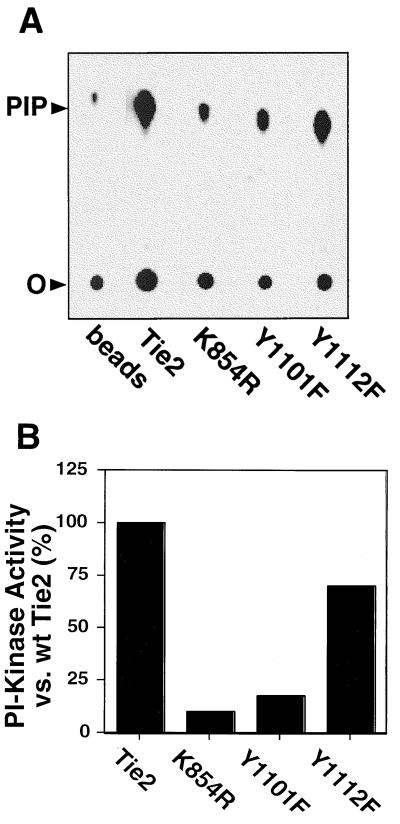
Having confirmed the specificity of the p85 interaction with Y1101, we evaluated whether p85 binding to Tie2 conferred association of functional lipid kinase activity by quantitatively measuring Tie2-associated PI3-kinase activity in vitro. As expected, the amount of receptor-associated PI3-kinase activity correlated with the ability of the wild-type or mutant Tie2 kinases to bind p85. Tie2-associated PI3-kinase activity was markedly reduced by both the K854R and Y1101F mutations (10 and 17% of wild-type Tie2-associated activity, respectively), whereas mutation of Y1112 to phenylalanine only modestly reduced receptor-associated activity (70% of wild-type level) (Fig. 4). These data demonstrate that p85 association with Tie2 confers functional PI3-kinase activity and support Y1101 as the preferred binding site for the PI3-kinase holoenzyme on Tie2.
FIG. 4.
Tie2-associated p85 confers association of PI3 kinase activity in vitro. (A) Representative thin-layer chromatogram demonstrating GST-Tie2 kinase-associated PI3-kinase activity. Autophosphorylated wild-type or mutant GST-kinase fusion proteins were immobilized on glutathione-Sepharose and incubated with endothelial cell lysates as described for Fig. 2 except that, after the final wash, protein complexes were incubated with PI and [γ-32P]ATP in kinase buffer. Radiolabeled PI3P was separated by thin-layer chromatography and visualized by autoradiography. As a control, glutathione-Sepharose without recombinant kinase was incubated with endothelial cell lysates (beads). The positions of the origin (O) and PI3P (PIP) are shown. (B) Quantitation of Tie2-associated PI3-kinase activity. PI3-kinase activity associated with wild-type or mutant GST-Tie2 from the chromatogram in panel A was quantitated with a Molecular Dynamics PhosphorImager. Activity was corrected for that associated with beads alone and expressed as a percentage of activity associated with the wild-type (wt) kinase.
An fms-Tie2 chimera is functionally active in NIH 3T3 cells.
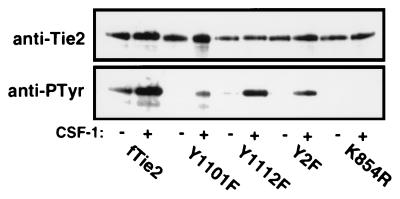
During the majority of these studies, an activating ligand for Tie2 was unknown. Therefore, to evaluate the p85-Tie2 association in vivo, we expressed a chimeric receptor consisting of the extracellular and transmembrane domains of c-fms, the CSF-1 receptor, and the cytoplasmic kinase domain of Tie2 (designated fTie2) in fibroblasts, with or without specific mutations. To demonstrate that wild-type and mutant fTie2 chimeric receptors were functionally active, cells expressing each receptor were evaluated for activation by CSF-1 in the presence of sodium orthovanadate in order to maximize receptor autophosphorylation. Importantly, the wild-type fTie2 and each of the tyrosine-to-phenylalanine mutants demonstrated CSF-1-dependent autophosphorylation, while the kinase-inactive K854R mutant remained unphosphorylated even after stimulation (Fig. 5). Consistent with results in vitro, the Shp2 binding site mutants (Y1112F and Y2F) were heavily tyrosine phosphorylated despite the loss of potentially important autophosphorylation sites (Fig. 5). In contrast, Y1101F was less phosphorylated than the wild-type kinase, possibly due to both the loss of an important autophosphorylation site and incomplete inhibition of Shp2 or other tyrosine phosphatases by vanadate (Fig. 5). However, these data demonstrate that mutation of Y1101 or Y1112, either individually or together, does not disrupt fTie2 kinase activity.
FIG. 5.
Chimeric fms-Tie2 receptors are functionally active in NIH 3T3 cells. NIH 3T3 fibroblasts stably expressing fms-Tie2 (fTie2) chimeric receptors, with or without specific mutations, were stimulated with CSF-1 (500 ng/ml) or left unstimulated, all in the presence of 1 mM sodium orthovanadate. Lysates were immunoprecipitated with an anti-c-fms MAb, separated by SDS–6% PAGE, and analyzed by Western blotting with anti-Tie2 and antiphosphotyrosine (anti-PTyr) antibodies to demonstrate CSF-1-dependent receptor autophosphorylation.
Tie2 induces plasma membrane translocation of a GFP-Akt PH domain fusion protein.
The fTie2-expressing cells were next used to evaluate the association of p85 with the Tie2 kinase in vivo. In spite of readily detectable association in yeast and in vitro, we were unable to demonstrate coimmunoprecipitation of p85 with fTie2 in any of these cell lines (data not shown). We were also unable to detect increases in PI3-kinase activity after receptor stimulation by conventional methods, such as in anti-fms or antiphosphotyrosine immunoprecipitates (data not shown). Therefore, we used a novel approach to evaluate PI3-kinase activation by Tie2 in vivo.
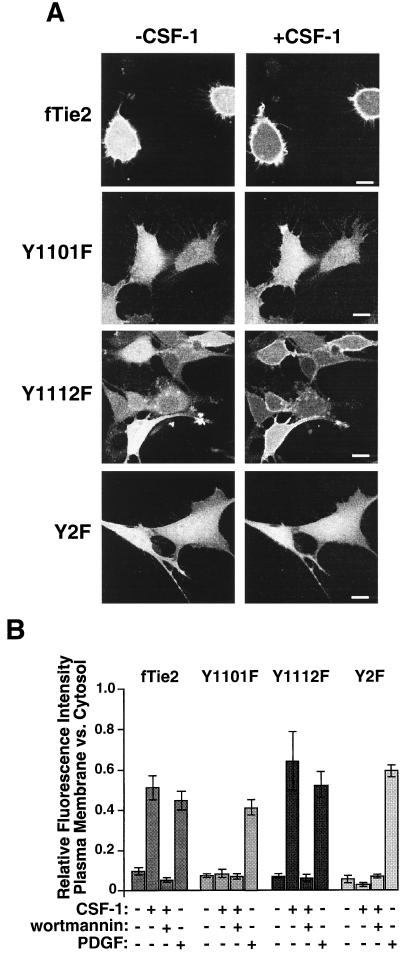
Because the PH domain of the Akt kinase is known to bind to the phospholipid products of activated PI3-kinase (33, 62), we used plasma membrane translocation of a GFP-Akt PH domain fusion protein (GFP-PH) as a surrogate marker for PI3-kinase activation in vivo. Stimulation of either the wild-type fTie2 or the Y1112F mutant resulted in a rapid (visible within 3 to 5 min) and reproducible plasma membrane localization of GFP-PH (Fig. 6) that was blocked by pretreatment with 50 nM wortmannin (Fig. 6B), demonstrating that the response was PI3-kinase dependent. Conversely, stimulation of either Y1101F or Y2F failed to induce membrane translocation of GFP-PH (Fig. 6). Stimulation of untransfected 3T3 cells with CSF-1 and vanadate or of fTie2 cells with vanadate alone produced no detectable response (data not shown), confirming that this effect was specific for the fTie2 chimera and not due to either another receptor or nonspecific effects of vanadate. Furthermore, stimulation of each cell line with PDGF induced plasma membrane localization of GFP-PH, demonstrating intact PI3-kinase signaling pathways in all of these cell lines (Fig. 6B). Together, these findings strongly suggest that fTie2 stimulation in NIH 3T3 fibroblasts results in PI3-kinase activation and a subsequent increase in membrane d-3-phosphoinositides. Furthermore, these results suggest that PI3-kinase activation by Tie2 is dependent upon association of p85 with Y1101 of Tie2.
FIG. 6.
fTie2 activation induces plasma membrane translocation of GFP-PH in vivo. (A) Representative images of wild-type or mutant fTie2 cells expressing GFP-PH, before and after stimulation with CSF-1. NIH 3T3 fibroblasts expressing fTie2 chimeric receptors, with or without specific mutations, were plated on coverslips, electroporated with RNA encoding GFP-PH, briefly serum starved, and then stimulated with CSF-1 (500 ng/ml) and vanadate (1 mM). Cells were analyzed by confocal laser scanning fluorescence microscopy before and after stimulation. Bar, 10 μm. (B) Quantitation of plasma membrane translocation of GFP-PH induced by either fTie2 or PDGFR stimulation. The relative fluorescence intensity of plasma membrane versus that of cytosol was assessed as described in Materials and Methods before and after stimulation with CSF-1 (500 ng/ml) or PDGF-BB (40 ng/ml), both in the presence of 1 mM sodium orthovanadate. Each cell line was also assessed after stimulation with CSF-1 and vanadate following pretreatment with 50 nM wortmannin. The relative fluorescence intensity in each experiment is expressed as the mean ± SEM from 10 to 20 cells.
Tie2 activates Akt in vivo.
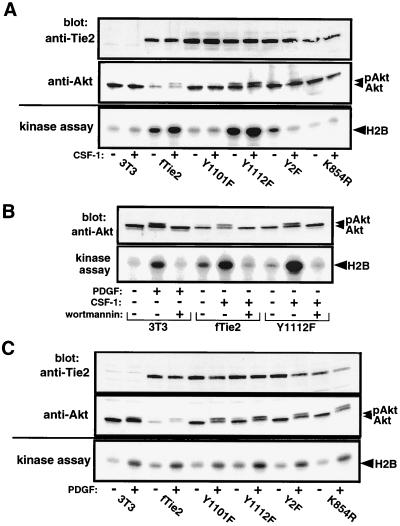
To investigate whether Tie2 activation of PI3-kinase results in activation of downstream signaling pathways, activation of Akt was evaluated by gel mobility shift on Western blots and in kinase assays with Akt immunoprecipitates. In both of these assays, results paralleled those of the GFP-PH experiments. Cell lines expressing fTie2 with an intact Y1101 residue (wild type and Y1112F) demonstrated an electrophoretic mobility shift of Akt after CSF-1 stimulation, consistent with the activated, phosphorylated form of the kinase (Fig. 7A, middle panel). Similarly, kinase assays demonstrated that Akt activity was increased by CSF-1 stimulation only in cells expressing fTie2 receptors with an intact p85 binding site (Fig. 7A, bottom panel). Untransfected 3T3 cells and those expressing the kinase-inactive mutant of fTie2 failed to show Akt activation after CSF-1 stimulation (Fig. 7A). Pretreatment of cells expressing wild-type fTie2 or the Y1112F mutant with 100 nM wortmannin prior to stimulation with CSF-1 abolished Akt phosphorylation and the increase in Akt kinase activity (Fig. 7B). Confirming the integrity of Akt and its upstream signaling pathways in all of these cell lines, PDGF induced both a mobility shift of Akt and an increase in Akt kinase activity in all cell lines tested (Fig. 7C). Taken together, these data demonstrate that Akt is activated in vivo by Tie2, and this activation requires both PI3-kinase and Tie2 kinase activity as well as an intact p85 binding site on Tie2.
FIG. 7.
Stimulation of fTie2 in vivo induces phosphorylation and activation of Akt. (A) Phosphorylation and activation of Akt requires an intact Y1101 residue on Tie2. Untransfected NIH 3T3 cells or those expressing fTie2 chimeric receptors were left unstimulated or stimulated with CSF-1 (500 ng/ml), all in the presence of 1 mM sodium orthovanadate. Aliquots of total cell lysates were separated by SDS–8% PAGE and blotted with anti-Tie2 PAb to demonstrate similar receptor expression levels (top) and anti-Akt PAb to evaluate Akt expression and demonstrate gel mobility shift secondary to phosphorylation (middle). Anti-Akt immunoprecipitates from the same cell lysates were used in kinase assays with the substrate histone H2B and [γ-32P]ATP. Proteins were separated by SDS–15% PAGE and evaluated by autoradiography to assess Akt activation after CSF-1 stimulation (bottom). (B) Tie2-mediated phosphorylation and activation of Akt is PI3-kinase dependent. Fibroblasts expressing wild-type fTie2 or Y1112F were either unstimulated, stimulated with CSF-1, or stimulated with CSF-1 following 15 min of pretreatment with 100 nM wortmannin, all in the presence of 1 mM vanadate. Untransfected 3T3 cells were treated similarly with PDGF-BB (30 ng/ml) as a positive control. Cell lysates were used to assess Akt phosphorylation by Western blotting and in kinase assays to evaluate Akt activation as described for panel A. (C) Wild-type- and mutant fTie2-expressing cells exhibit intact Akt signaling pathways. Untransfected 3T3 cells and cells expressing wild-type or mutant fTie2 receptors were evaluated as described for panel A, except that stimulation of all cells was with PDGF (30 ng/ml) to demonstrate intact signaling pathways upstream of Akt. The positions of Akt, phosphorylated Akt (pAkt), and histone H2B are shown.
Tie2 activation in vivo induces an increase in PI3P and PI 3,4-bisphosphate [PI(3,4)P2].
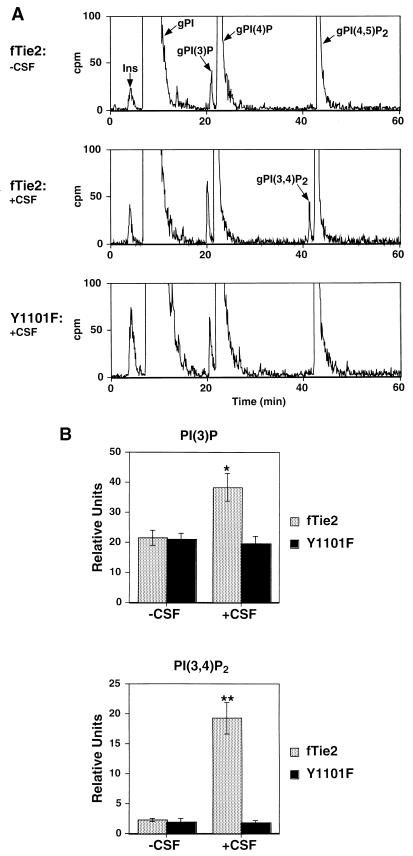
Results from the GFP-PH experiments and analysis of Akt activation demonstrated that Tie2 activation of PI3-kinase requires an intact Y1101 residue. To verify that these results correlated with increases in d-3-phosphoinositides in vivo, we measured PI phosphates in cells expressing either wild-type fTie2 or the Y1101F mutant by HPLC analysis of extracts from cells labeled to steady state with [3H]myo-inositol. When fTie2 cells were stimulated with CSF-1, a significant increase was seen in PI3P and, more importantly, PI(3,4)P2, which has been shown to bind and activate Akt (33) (Fig. 8). In contrast, Y1101F cells stimulated with CSF-1 failed to show any change in either of these phospholipids (Fig. 8). No PI 3,4,5-trisphosphate (PIP3) was detected in either cell line, possibly due to insufficient sensitivity in detecting this lipid product by steady-state labeling of phosphoinositides with [3H]inositol, and no changes in d-4-phosphoinositides were noted (data not shown). These findings confirm that fTie2 activates PI3-kinase in vivo and that this effect requires an intact Y1101 residue on Tie2. Tie2 activation induces a measurable, significant increase in cellular PI(3,4)P2 production that may account for downstream activation of Akt. Notably, these results also provide support for using plasma membrane translocation of a GFP-Akt PH domain fusion protein as a surrogate marker of PI3-kinase activation.
FIG. 8.
fTie2 activation of PI3-kinase and increases in cellular PI3P and PI 3,4-bisphosphate require an intact Y1101 residue on Tie2. (A) Representative chromatograms from wild-type fTie2- and Y1101F-expressing cells. Cells labeled for 72 h with 5 μCi of [3H]myo-inositol per ml were serum starved for 2 h and then were either left unstimulated or stimulated with CSF-1, all in the presence of 1 mM vanadate. Cellular lipids were extracted and deacylated, and then the resultant gPIPs were resolved by HPLC as described in Materials and Methods. Positions of the different gPIPs are indicated. cpm, counts per minute; Ins, free inositol. (B) Quantitative analysis of d-3-phosphoinositide production by wild-type fTie2 and Y1101F. Estimated baseline counts per minute on each chromatogram were subtracted from total counts under each peak and then expressed relative to the total cellular gPI in each experiment. Each bar represents the mean ± SEM from three separate experiments. ∗, P < 0.05 versus fTie2 minus CSF; P < 0.02 versus Y1101F plus CSF. ∗∗, P = 0.02 versus fTie2 minus CSF; P < 0.03 versus Y1101F plus CSF.
DISCUSSION
In this report, we have demonstrated that p85 associates with tyrosine 1101 of the activated Tie2 kinase. Tie2 was found to activate PI3-kinase in vivo as demonstrated by direct measurement of increases in cellular PI3Ps and by plasma membrane translocation of a GFP-Akt PH domain fusion protein. Furthermore, Tie2 activation in vivo resulted in downstream activation of Akt in a PI3-kinase-dependent manner. Based on what is known about the cellular functions of PI3-kinase and Akt, these findings may have important implications regarding the role of Tie2 signaling in both the embryonic and the adult vasculature.
Although the results from the yeast two-hybrid system and in vitro association assays clearly demonstrated that p85 is capable of associating with Tie2, it remains unclear whether this interaction is direct or is mediated by an adapter protein, such as Grb2 or Gab1. Based on its similar binding site preference for Y1101, Grb2 seems a logical candidate adapter for the p85-Tie2 interaction. In fact, a direct interaction between the SH3 domain of Grb2 and the bcr region of p85 has been demonstrated previously (67). Similarly, Gab1 has been shown to bind to YVNX motifs such as that at Y1101 (46), and this adapter molecule is responsible for the association of p85 with other activated RTKs (28, 29, 46). However, the identification of p85 as an interactor for Tie2 by the yeast two-hybrid system supports a direct interaction. In rare cases, bridging proteins have been shown to be responsible for two-hybrid interactions (25), but because they occur in the yeast nucleus, these intermediates are likely to be yeast transcription factors. Thus, while interaction via an adapter protein cannot be ruled out, the simplest interpretation of our results is that p85 associates directly with Tie2 at tyrosine 1101.
Analysis of the amino acid residues flanking Y1101 demonstrates that this residue does not match the consensus binding motif for p85 (58); rather, it closely resembles a number of other nonconsensus p85 binding proteins that lack the preferred methionine in the Y+3 position (5, 7, 8, 50) (Fig. 9). Notably, Tie2 differs from all of these proteins by the presence of a threonine in this position (T1104). The crystal structure of the N-terminal SH2 domain of p85 demonstrates a hydrophobic pocket for the side chains of the Y+1 and Y+3 residues (48); the primary structure of the C-terminal SH2 domain predicts a similar tertiary structure. Thus, T1104 may dictate a lower affinity or a more transient interaction between p85 and Tie2 and may account for difficulty in detecting association in vivo. Consistent with the possibility that binding of p85 to the nonconsensus motif on Tie2 is of lower affinity, coimmunoprecipitation of p85 with the consensus site on the PDGF receptor (PDGFR) is readily detectable (data not shown). Since Tie2 appears, at least qualitatively, to activate both PI3-kinase and Akt as well as does the PDGFR in the same cells (Fig. 6B and 7B), an important implication of these findings is that even a more transient or lower-affinity p85-receptor interaction can result in membrane recruitment and activation of PI3-kinase.
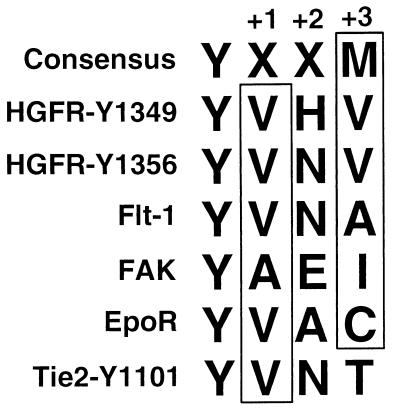
FIG. 9.
Comparison of amino acid residues flanking tyrosine 1101 of Tie2 with consensus and nonconsensus p85 binding motifs. Two nonconsensus p85 binding motifs on the hepatocyte growth factor receptor (HGFR) as well as those on the endothelium-specific vascular endothelial growth factor receptor Flt-1, focal adhesion kinase (FAK), and the erythropoietin receptor (EpoR) are shown in comparison to residues flanking Y1101 of Tie2 and the consensus motif determined from phosphopeptide binding studies (58). There is a strong preference for hydrophobic or neutral residues in the Y+1 and Y+3 positions (boxed residues). Y1101 of Tie2 differs by the presence of a threonine in the Y+3 position. Amino acid residues are designated according to the single-letter code: A, alanine; C, cysteine; E, glutamate; H, histidine; I, isoleucine; M, methionine; N, glutamine; V, valine; Y, tyrosine; X, any amino acid residue.
Activation of PI3-kinase is believed to involve both receptor binding and membrane localization (34, 54), although the latter has been shown to be sufficient for activation of a number of downstream signaling pathways (34). Recent evidence has demonstrated that one product of PI3-kinase activation, PIP3, can bind to the SH2 domains of p85 and is able to compete with tyrosyl phosphoproteins, including phosphorylated RTKs, for p85 binding (53). This property of PI3-kinase might explain the difficulty in demonstrating p85-Tie2 association by coimmunoprecipitation and the lack of PI3-kinase activity in anti-fms or antiphosphotyrosine immunoprecipitates. More importantly, it provides evidence for a positive feedback loop for PI3-kinase activation. Recruitment of a threshold amount of PI3-kinase to the cell membrane might generate enough PIP3 to recruit more PI3-kinase, which could potentiate this response. Increased PIP3 might then recruit other SH2- and PH domain-containing proteins to the plasma membrane. Indeed, phospholipase Cγ has recently been shown to be activated by binding of its SH2 domains to PIP3 (3). Similarly, activation of both Akt and p70 S6 kinase is now known to be mediated in part by recruitment of the 3-phosphoinositide-dependent kinases PDK1 and PDK2 to the plasma membrane by binding of their PH domains to PIP3 (51, 61). These findings might help explain how even a transient p85-Tie2 interaction could generate amplified downstream signals to create a localized signaling domain at the plasma membrane.
Debate exists regarding which phospholipid products of PI3- kinase are responsible for mediating downstream signaling events. In our experiments, increases in PI(3,4)P2 in the fTie2 cells correlated well with GFP-PH translocation and with phosphorylation and activation of Akt (Fig. 6 and 7). These findings agree with those of Klippel et al. (33) demonstrating that PI(3,4)P2, but not PI3P or PIP3, can activate Akt. In contrast, other investigators have suggested that PIP3 is involved in Akt activation (62). Notably, our analysis of phospholipid products generated by fTie2 activation failed to detect PIP3. One explanation for this finding is that steady-state labeling of lipids with [3H]inositol is not sufficiently sensitive to detect what may be relatively small amounts of PIP3 produced in these cells. This problem may be circumvented by using [3H]inositol of higher specific activity, by assessing PIP3 production at different times after Tie2 activation, or by pulse-labeling cells with 32Pi. A more intriguing explanation for this finding is provided by the recent identification of the SH2 domain-containing inositol 5-phosphatase SHIP (40). Tie2 activation might result in the generation of PIP3 and at the same time recruit binding of SHIP. SHIP could then dephosphorylate PIP3 to produce PI(3,4)P2, which appears to be a better activator of Akt (33). It is not yet clear whether SHIP is expressed either in endothelial cells or in the fibroblasts used in these studies, but it is interesting to note that SHIP can associate with Shp2 (41), which we have also shown to bind to Tie2 (31) and which is capable of dephosphorylating the activated Tie2 receptor (35). Thus, regulation of Tie2 signaling might involve a complex interplay of a number of signaling proteins and phospholipids.
Activation of PI3-kinase by a variety of signaling pathways has been shown to result in a wide range of cellular responses, including mitogenesis, regulation of the cytoskeleton, and cell survival (reviewed in reference 4). It is not clear how receptor specificity for these individual responses is determined, but PI3-kinase has been shown to transduce its signals via a number of potential downstream effectors, including MAP kinase, p70 S6 kinase, c-Jun N-terminal kinase, and Akt (34). Recently, a number of studies have shed light on the role of PI3-kinase-dependent activation of Akt with the demonstration that this process prevents apoptosis (1, 9, 13, 38). This phenomenon appears in part to be due to serine phosphorylation of the cell death protein BAD by Akt, which prevents heterodimerization of BAD with the antiapoptotic proteins Bcl-2 and Bcl-xL (9, 11). Phosphorylated BAD is then thought to be inactivated by sequestration at the cell membrane by 14-3-3 proteins (9). Whether Akt phosphorylates BAD in response to Tie2 activation of PI3-kinase in endothelial cells is not yet known, but this possibility is the focus of ongoing experiments.
Based on recent genetic studies of Tie2 and its ligands, Ang1 and Ang2 (15, 43, 55, 64), Tie2 is thought to be important in the maturation and maintenance of the adult vasculature (21, 24). Although still not well understood, these endothelial cell processes may involve a variety of cellular functions, including cytoskeletal integrity, cell-cell or cell-matrix interactions, and cell proliferation and/or survival. Considering the varied functions attributed to PI3-kinase, it is possible that Tie2 activation of PI3-kinase could be involved in multiple aspects of these processes. Together with the results presented here, data linking Akt to antiapoptosis raise the possibility that Tie2 could be involved in endothelial cell survival. As the endogenous Tie2 ligand, Ang1, becomes available, future studies should be able to address the question of whether PI3-kinase and Akt are activated by Tie2 in endothelial cells, and studies of endothelial cell survival will be critical to evaluate the function of this potentially important signaling cascade downstream of Tie2. Further understanding of these and other signaling pathways activated by Tie2 will likely provide significant insights into the role of this novel receptor in endothelial biology.
ACKNOWLEDGMENTS
We acknowledge Anke Klippel for generously providing the p85 antibodies and for advice regarding the PI3-kinase and Akt kinase assays, Shuling Guo for assistance with the lipid deacylations and HPLC analysis, Roger Brent for providing the yeast strains and yeast two-hybrid plasmids, Arthur Bank for providing GP+E86 packaging cells, and Genetics Institute for generously supplying CSF-1.
This work was supported in part by grants from the National Institutes of Health (HL 55265 to K.G.P., HL 03557 to C.D.K., GM 51457 to T.M., and HL 55672 to J.D.Y.), from the James S. McDonnell Foundation (to K.G.P.), and from Procter & Gamble (grant SR 1617 to T.M.). C.D.K. was supported in part by a Fellowship Award from the North Carolina Affiliate of the American Heart Association. T.P.S. is a recipient of a fellowship from the Swiss National Science Foundation (grant 823A-050457). W.-P.Y. and M.A.B. were supported by the Bristol-Myers Squibb Pharmaceutical Research Institute. J.D.Y. is the recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences, and T.M. was supported by a fellowship from the David and Lucille Packard Foundation.
REFERENCES
- 1.Ahmed N N, Grimes H L, Bellacosa A, Chan T O, Tsichlis P N. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonetti D A, Algenstaedt P, Kahn C R. Insulin receptor substrate 1 binds two novel splice variants of the regulatory subunit of phosphatidylinositol 3-kinase in muscle and brain. Mol Cell Biol. 1996;16:2195–2203. doi: 10.1128/mcb.16.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae Y S, Cantley L G, Chen C-S, Kim S-R, Kwon K-S, Rhee S G. Activation of phospholipase C-γ by phosphatidylinositol 3,4,5-triphosphate. J Biol Chem. 1998;273:4465–4469. doi: 10.1074/jbc.273.8.4465. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter C L, Cantley L C. Phosphoinositide kinases. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen H-C, Appeddu P A, Isoda H, Guan J-L. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. J Biol Chem. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- 6.Crameri A, Whitehorn E A, Tate E, Stemmer P C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham S A, Waxham M N, Arrate P M, Brock T A. Interaction of the Flt-1 tyrosine kinase receptor with the p85 subunit of phosphatidylinositol 3-kinase: mapping of a novel site involved in binding. J Biol Chem. 1995;270:20254–20257. doi: 10.1074/jbc.270.35.20254. [DOI] [PubMed] [Google Scholar]
- 8.Damen J E, Cutler R L, Jiao H, Yi T, Krystal G. Phosphorylation of tyrosine 503 in the erythropoietin receptor (EpR) is essential for binding the p85 subunit of phosphatidylinositol (PI) 3-kinase and for EpR-associated PI 3-kinase activity. J Biol Chem. 1995;270:23402–23408. doi: 10.1074/jbc.270.40.23402. [DOI] [PubMed] [Google Scholar]
- 9.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 10.Davis S, Aldrich T H, Jones P F, Acheson A, Compton D L, Jain V, Ryan T E, Bruno J, Radziejewski C, Maisonpierre P C, Yancopoulos G D. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 11.del Peso L, González-García M, Page C, Herrera R, Nuñez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 12.Dubois N A, Kolpack L C, Wang R, Azizkhan R G, Bautch V L. Isolation and characterization of an established endothelial cell line from transgenic mouse hemangiomas. Exp Cell Res. 1991;196:302–313. doi: 10.1016/0014-4827(91)90265-v. [DOI] [PubMed] [Google Scholar]
- 13.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R S, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 14.Dumont D J, Fong G-H, Gradwohl G, Alitalo K, Breitman M L. Vascularization of the mouse embryo: a study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- 15.Dumont D J, Gradwohl G, Fong G-H, Puri M C, Gertsenstein M, Auerbach A, Breitman M L. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 16.Dumont D J, Gradwohl G J, Fong G-H, Auerbach R, Breitman M L. The endothelial-specific receptor tyrosine kinase, tek, is a member of a new subfamily of receptors. Oncogene. 1993;8:1293–1301. [PubMed] [Google Scholar]
- 17.Dumont D J, Yamaguchi T P, Conlon R A, Rossant J, Breitman M L. tek, a novel tyrosine kinase gene located on mouse chromosome 4, is expressed in endothelial cells and their presumptive precursors. Oncogene. 1992;7:1471–1480. [PubMed] [Google Scholar]
- 18.Edgell C J S, McDonald C C, Graham J B. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc Natl Acad Sci USA. 1983;80:3734–3737. doi: 10.1073/pnas.80.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fantl W J, Johnson D E, Williams L T. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 21.Folkman J, D’Amore P A. Blood vessel formation: what is its molecular basis? Cell. 1996;87:1153–1155. doi: 10.1016/s0092-8674(00)81810-3. [DOI] [PubMed] [Google Scholar]
- 22.Golemis E A, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify protein interactions. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. pp. 13.14.1–13.14.17. [Google Scholar]
- 23.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D. Signaling vascular morphogenesis and maintenance. Science. 1997;277:48–50. doi: 10.1126/science.277.5322.48. [DOI] [PubMed] [Google Scholar]
- 25.Hannon G J, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 26.Heim R, Cubitt A B, Tsien R. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 27.Hiles I D, Otsu M, Volinia S, Fry M J, Gout I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty N F, Hsuan J J, Courtneidge S A, Parker P J, Waterfield M D. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell. 1992;70:419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 28.Holgado-Madruga M, Emlet D R, Moscatello D K, Godwin A K, Wong A J. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 29.Holgado-Madruga M, Moscatello D K, Emlet D R, Dieterich R, Wong A J. Grb2-associated binder-1 mediates phosphatidylinositol 3-kinase activation and the promotion of cell survival by nerve growth factor. Proc Natl Acad Sci USA. 1997;94:12419–12424. doi: 10.1073/pnas.94.23.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holt K H, Olson A L, Moye-Rowley W S, Pessin J E. Phosphatidylinositol 3-kinase activation is mediated by high-affinity interactions between distinct domains within the p110 and p85 subunits. Mol Cell Biol. 1994;14:42–49. doi: 10.1128/mcb.14.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Turck C W, Rao P, Peters K G. GRB2 and SH-PTP2: potentially important endothelial signaling molecules downstream of the TEK/TIE2 receptor tyrosine kinase. Oncogene. 1995;11:2097–2103. [PubMed] [Google Scholar]
- 32.Klippel A, Escobedo J A, Hirano M, Williams L T. The interaction of small domains between the subunits of phosphatidylinositol 3-kinase determines enzyme activity. Mol Cell Biol. 1994;14:2675–2685. doi: 10.1128/mcb.14.4.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klippel A, Kavanaugh W M, Pot D, Williams L T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klippel A, Reinhard C, Kavanaugh W M, Appel G, Escobedo M-A, Williams L T. Membrane localization of phosphatidylinositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Kontos C D, Huang L, Yang W-P, Blanar M, Peters K G. The p85 subunit of phosphatidylinositol 3′-kinase associates with the endothelium-specific receptor tyrosine kinase Tek. Circulation. 1996;94:I–405. [Google Scholar]
- 35.Kontos, C. D., L. Huang, and K. G. Peters. 1998. Unpublished data.
- 36.Kontos, C. D., and K. G. Peters. Unpublished data.
- 37.Korhonen J, Polvi A, Partanen J, Alitalo K. The mouse tie receptor tyrosine kinase gene: expression during embryonic angiogenesis. Oncogene. 1994;9:395–403. [PubMed] [Google Scholar]
- 38.Kulik G, Klippel A, Weber M J. Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin P, Polverini P, Dewhirst M, Shan S, Rao P S, Peters K. Inhibition of tumor angiogenesis using a soluble receptor establishes a role for Tie2 in pathologic vascular growth. J Clin Invest. 1997;100:2072–2078. doi: 10.1172/JCI119740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Damen J E, Ware M, Hughes M, Krystal G. SHIP, a new player in cytokine-induced signalling. Leukemia. 1997;11:181–184. doi: 10.1038/sj.leu.2400559. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Damen J E, Ware M D, Krystal G. Interleukin-3 induces the association of the inositol 5-phosphatase SHIP with SHP2. J Biol Chem. 1997;272:10998–11001. doi: 10.1074/jbc.272.17.10998. [DOI] [PubMed] [Google Scholar]
- 42.Maisonpierre P C, Goldfarb M, Yancopoulos G D, Gao G. Distinct rat genes with related profiles of expression define a TIE receptor tyrosine kinase family. Oncogene. 1993;8:1631–1637. [PubMed] [Google Scholar]
- 43.Maisonpierre P C, Suri C, Jones P F, Bartunkova S, Wiegand S J, Radziejewski C, Compton D, McClain J, Aldrich T H, Papadopoulos N, Daly T J, Davis S, Sato T N, Yancopoulos G D. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 44.Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen L, Holgado-Madruga M, Maroun C, Fixman E D, Kamikura D, Fournier T, Charest A, Tremblay M L, Wong A J, Park M. Association of the multisubstrate docking protein Gab1 with the hepatocyte growth factor receptor requires a functional Grb2 binding site involving tyrosine 1356. J Biol Chem. 1997;272:20811–20819. doi: 10.1074/jbc.272.33.20811. [DOI] [PubMed] [Google Scholar]
- 47.Nishimura R, Li W, Kashishian A, Mondino A, Zhou M, Cooper J, Schlessinger J. Two signaling molecules share a phosphotyrosine-containing binding site in the platelet-derived growth factor receptor. Mol Cell Biol. 1993;13:6889–6896. doi: 10.1128/mcb.13.11.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolte R T, Eck E J, Schlessinger J, Shoelson S E, Harrison S C. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat Struct Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 49.Peters K G, Coogan A, Berry D, Marks J, Iglehart J D, Kontos C D, Rao P, Sankar S, Trogan E. Expression of Tie2/Tek in breast tumour vasculature provides a new marker for evaluation of tumour angiogenesis. Br J Cancer. 1998;77:51–56. doi: 10.1038/bjc.1998.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ponzetto C, Bardelli A, Maina F, Longati P, Panayotou G, Dhand R, Waterfield M, Comoglio P M. A novel recognition motif for phosphatidylinositol 3-kinase binding mediates its association with the hepatocyte growth factor/scatter factor receptor. Mol Cell Biol. 1993;13:4600–4608. doi: 10.1128/mcb.13.8.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 52.Puri M C, Rossant J, Alitalo K, Bernstein A, Partanen J. The receptor tyrosine kinase TIE is required for integrity and survival of vascular endothelial cells. EMBO J. 1995;14:5884–5891. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rameh L E, Chen C-S, Cantley L C. Phosphatidylinositol (3,4,5)P3 interacts with SH2 domains and modulates PI 3-kinase association with tyrosine-phosphorylated proteins. Cell. 1995;83:821–830. doi: 10.1016/0092-8674(95)90195-7. [DOI] [PubMed] [Google Scholar]
- 54.Rordorf-Nikolic T, Van Horn D J, Chen D, White M F, Backer J M. Regulation of phosphatidylinositol 3′-kinase by tyrosyl phosphoproteins: full activation requires occupancy of both SH2 domains in the 85-kDa regulatory subunit. J Biol Chem. 1995;270:3662–3666. doi: 10.1074/jbc.270.8.3662. [DOI] [PubMed] [Google Scholar]
- 55.Sato T N, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 56.Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 57.Skolnik E Y, Margolis B, Mohammadi M, Lowenstein E, Fischer R, Drepps A, Ullrich A, Schlessinger J. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 58.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 59.Stauffer T P, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane phosphatidylinositol-4,5 bisphosphate concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 60.Stauffer T P, Meyer T. Compartmentalized IgE-receptor-mediated signal transduction in living cells. J Cell Biol. 1997;139:1447–1454. doi: 10.1083/jcb.139.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter G F, Holmes A B, Gaffney P R J, Reese C B, McCormick F, Tempst P, Coadwell J, Hawkins P T. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-triphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 62.Stokoe D, Stephens L R, Copeland T, Gaffney P R J, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Dual role of phosphatidylinositol-3,4,5-triphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 63.Stolz L E, Kuo W J, Longchamps J, Sekhon M K, York J D. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- 64.Suri C, Jones P F, Patan S, Bartunkova S, Maisonpierre P C, Davis S, Sato T N, Yancopoulos G D. Requisite role for angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 65.Teruel M N, Meyer T. Electroporation induced formation of individual calcium entry sites in cell body and processes of adherent cells. Biophys J. 1997;73:1785–1796. doi: 10.1016/S0006-3495(97)78209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Geer P, Hunter T, Lindberg R A. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Auger K R, Jarvis L, Shi Y, Roberts T M. Direct association of Grb2 with the p85 subunit of phosphatidylinositol 3-kinase. J Biol Chem. 1995;270:12774–12780. doi: 10.1074/jbc.270.21.12774. [DOI] [PubMed] [Google Scholar]
- 68.Whitman M, Downes C P, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 69.Wong A L, Haroon Z A, Werner S, Dewhirst M W, Greenberg C S, Peters K G. Tie2 expression and phosphorylation in angiogenic and quiescent adult tissues. Circ Res. 1997;81:567–574. doi: 10.1161/01.res.81.4.567. [DOI] [PubMed] [Google Scholar]
- 70.Yokoe H, Meyer T. Spatial dynamics of GFP-tagged proteins investigated by local fluorescence enhancement. Nat Biotechnol. 1996;14:1252–1256. doi: 10.1038/nbt1096-1252. [DOI] [PubMed] [Google Scholar]
- 71.York J D, Majerus P W. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J Biol Chem. 1994;269:7847–7850. [PubMed] [Google Scholar]
- 72.Ziegler S F, Bird T A, Schneringer J A, Schooley K A, Baum P A. Molecular cloning and characterization of a novel receptor protein tyrosine kinase from human placenta. Oncogene. 1993;8:663–670. [PubMed] [Google Scholar]