Abstract
Large deletions of the upstream U3 sequences in the long terminal repeats (LTRs) of human immunodeficiency virus and simian immunodeficiency virus (SIV) accumulate in vivo in the absence of an intact nef gene. In the SIV U3 region, about 65 bp just upstream of the single NF-κB binding site always remained intact, and some evidence for a novel enhancer element in this region exists. We analyzed the transcriptional and replicative capacities of SIVmac239 mutants containing deletions or mutations in these upstream U3 sequences and/or the NF-κB and Sp1 binding sites. Even in the absence of 400 bp of upstream U3 sequences, the NF-κB site and all four Sp1 binding sites, the SIV promoter maintained about 15% of the wild-type LTR activity and was fully responsive to Tat activation in transient reporter assays. The effects of these deletions on virus production after transfection of COS-1 cells with full-length proviral constructs were much greater. Deletion of the upstream U3 sequences had no significant influence on viral replication when either the single NF-κB site or the Sp1 binding sites were intact. In contrast, the 26 bp of sequence located immediately upstream of the NF-κB site was essential for efficient replication when all core enhancer elements were deleted. A purine-rich site in this region binds specifically to the transcription factor Elf-1, a member of the ets proto-oncogene-encoded family. Our results indicate a high degree of functional redundancy in the SIVmac U3 region. Furthermore, we defined a novel regulatory element located immediately upstream of the NF-κB binding site that allows efficient viral replication in the absence of the entire core enhancer region.
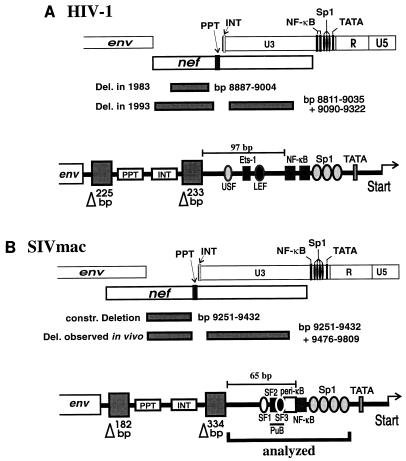
The proviral genomes of all retroviruses are flanked by repetitive sequences called long terminal repeats (LTRs) (38). Each LTR consists of three regions, U3, R, and U5 (38). The U3 region of the primate lentiviruses contains (i) sequences required for integration into the host cell genome and (ii) major transcriptional control elements: the TATA box motif and binding sites for the transcription factors NF-κB and Sp1 (9, 10, 12, 17, 18, 28, 34, 36). The U3 regions of human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) are considerably longer than those of other lentiviruses, ranging from about 450 to 560 bp in length (27). About 330 to 400 bp of the sequences upstream of the NF-κB and Sp1 binding sites overlap the nef open reading frame. After infection of rhesus macaques with a nef-defective SIV variant, large deletions accumulated within a 334-bp region of U3 that overlaps the nef open reading frame (22) (Fig. 1). Further analysis confirmed that this region serves predominantly as the Nef coding region (13). Similar deletions were observed in infection with nef-defective HIV-1 (4, 23) (Fig. 1). In one of these long-term nonprogressors of HIV-1 infection, the predominant deletion observed early in infection was located in the nef-unique region, and long deletions in U3 accumulated over time (23). These results suggest that in the absence of an intact nef gene, these upstream U3 sequences (US sequences) are lost and may not be advantageous for SIVmac and HIV-1 replication in vivo.
FIG. 1.
Locations of deletions in the nef-unique and U3 regions observed in HIV-1 and SIV infection. (A) Deletions observed in a long-term nonprogressor of HIV-1 infection at early and late time points (23). (B) Additional deletions observed in the U3 region in macaques infected with nef-deleted SIVmac239 (22). Nucleotide numbers refer to the HIV-1 NL4-3 (27) and SIVmac239 (32) sequences. Below, a schematic presentation of sequences close to the 3′ end that were maintained is given. The shaded boxes labeled with triangles indicate the deletions observed in the nef-unique and U3 regions in a long-term nonprogressor of HIV-1 infection (23) and rhesus macaques experimentally infected with nef-deleted SIVmac239 (22). As indicated, sequence elements of well-documented functional relevance, e.g., the polypurine tract, the core enhancer, and the TATA box, were preserved. Abbreviations: INT, U3 sequences required for integration; PPT, polypurine tract; USF, upstream stimulatory factor; Ets, Ets family transcription factor; SF, simian factor (40).
These naturally occurring deletions (NOD) removed up to 65% of the HIV-1 and SIVmac U3 regions and almost the entire nef-unique region. However, some elements of well-documented importance, like the polypurine tract, the NF-κB and Sp1 binding sites, the TATA box motif, and sequences required for proviral integration, were not affected (Fig. 1) (22, 23). In both HIV-1 and SIVmac U3, also 60 to 90 bp just upstream of the NF-κB binding site(s) were always preserved (22, 23). For HIV-1, it has been documented that the cell-type-specific cellular transcription factors USF, LEF-1, and Ets-1 bind to this region (Fig. 1) and activate transcription synergistically with Sp1 and NF-κB on chromatin-assembled DNA (1, 6, 20, 30, 35). Furthermore, point mutations in these binding sites decreased viral replication (21).
Relatively little is known about the relevance of the sequences just upstream of the single NF-κB binding site in the SIVmac LTR for viral transcription and replication. Several lines of evidence suggest the presence of an important enhancer element in this region. For instance, some transcription factors that bind just upstream of the NF-κB binding site in the LTRs of SIVmac (33, 40) and the closely related HIV-2 have been described (24–26). Most strikingly, SIVmac239 containing deletions of all NF-κB and Sp1 binding sites replicated with kinetics similar to those of the wild-type SIVmac239 clone in lymphoid cells and caused AIDS in rhesus macaques (14, 15). In contrast, HIV-1 containing deletions or substitutions in all NF-κB and Sp1 elements is not competent for replication (34).
In this study, we have analyzed the influence of deletions and point mutations in the region upstream of the core enhancer region. Since 334 bp of US sequences are clearly dispensable for SIV LTR function in vivo (22), we focused on the 65 bp just upstream of the NF-κB binding site, designated the US65 region (Fig. 2). Deletion of this region had only a moderately negative effect on transcriptional activity in transient assays and did not reduce viral replication in several cell lines, including primary rhesus macaque peripheral blood mononuclear cells (rPBMC), when either the single NF-κB binding site or the four Sp1 binding sites were present. Therefore, we analyzed SIVmac239 LTR variants containing deletions in the US sequences in conjunction with deletion of the entire core enhancer. We report that a purine-rich site (purine-rich box 2 [PuB2]) in the 26 bp of sequences located just upstream of the NF-κB binding site binds to the transcription factor Elf-1 and that this regulatory element allows efficient viral replication in the absence of the entire core enhancer region.
FIG. 2.
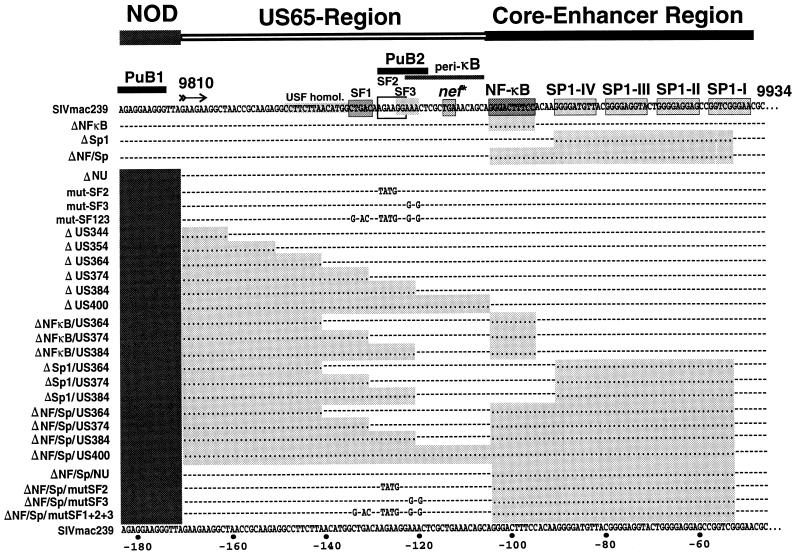
Mutations in the SIVmac239 U3 region. The SIVmac239 sequence has been published by Regier and Desrosiers (32). The NF-κB and Sp1 binding sites, the stop codon of nef, two PuB sites, a region with homology to the USF binding site, the peri-κB binding site (2), and the SF1 to SF3 sites (40) are indicated. Dots indicate deletions; dashes indicate identities in sequence. The deleted areas are shaded for clarity.
MATERIALS AND METHODS
Construction of LTR mutants.
An overview of the LTR mutants analyzed is given in Fig. 2. LTR mutations were introduced into the proviral clone SIVmac239 ΔNU, in which 182 bp in the nef-unique region and 334 bp of the upstream U3 region are deleted (11). The 334-bp U3 deletion affects only sequences that were selectively deleted in macaques infected with nef-defective SIVmac239 (22). First, site-directed mutagenesis was performed by spliced overlap extension PCR to generate unique SstII and XhoI sites at the 5′ end of the US65 region (ΔUSK-mutant). PCR products were gel purified and inserted into the SIVmac239 ΔNU clone by using the unique XmaI and EcoRI sites in the nef-unique region and in the vector sequences flanking the 3′ end of the provirus. Subsequently, the mutated 3′ LTR was amplified with primers pSphI (5′-CATTAAAGCTTGTCGACTGGAAGGGATTTAT-3′) and pSphII (5′-AATTGGCGCCAATCTGCTAGGGATTTTCCTG-3′) and inserted into the HindIII site located upstream of the 5′ end of the provirus and the NarI site located at the 3′ end of the 5′ LTR. Cloning of this fragment generated a unique SalI site upstream of the 5′ end of the provirus. All point mutants (simian factor 2 [SF2], SF3, and SF123 binding sites) and most deletion mutants were also generated by spliced overlap extension PCR. Mutations were first introduced into the 3′ LTR. Subsequently, the mutated 3′ LTRs were PCR amplified and inserted into the unique SalI and NarI sites, to replace the 5′ LTR. Mutations were always introduced into both LTRs to prevent recombination. SIVmac239 LTR mutants containing deletions in the upstream U3 region in conjunction with deletions in the core enhancer were generated by using the same protocol except that templates with deletions in the NF-κB and/or Sp1 binding sites were used in the first round of PCR. The ΔUS400 mutant was created by using a 5′ primer containing the XmaI site, the polypurine tract, and the NF-κB binding site. All PCR-derived inserts were sequenced to confirm that only the intended changes were present.
Transactivation assays.
The mutated LTRs (Fig. 2) were PCR amplified and cloned upstream of the luciferase reporter gene. For Tat expression, the first coding exon of the SIVmac239 tat gene was cloned into an expression plasmid containing a cytomegalovirus promoter (pcDNAIII; Invitrogen, San Diego, Calif.). To assess the promoter activities, COS-1 cells were seeded at a fivefold dilution and transfected with 1 μg of the various SIVmac LTR-luciferase plasmids, or cotransfected with 1 μg of a SIVmac Tat expression plasmid, by a DEAE-dextran method on the following day (3). The cells were harvested 48 h after transfection, and luciferase activity was measured with a commercially available kit as recommended by the manufacturer (Promega, Madison, Wis.). To allow a better comparison among individual experiments, stock DNA preparations from an SIVmac239 wild-type (239wt) LTR control plasmid were always included as a positive control. The amount of luciferase activity obtained after transfection of 239wt LTR plasmid was considered 100% activity, and the activities of the mutated LTRs were determined relative to this value. Furthermore, 1 μg of a plasmid expressing the β-galactosidase gene under control of the cytomegalovirus promoter was cotransfected as an internal control for transfection efficiency. The luciferase values were normalized for protein content and β-galactosidase activity, which were assayed by using commercially available kits (Promega).
Production of virus stocks.
For virus production, semiconfluent COS-1 cells were transfected with 5 μg of full-length proviral DNA constructs as described above. At 48 h posttransfection, the medium was replaced with fresh Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS). Virus stocks were harvested at 4 days posttransfection from cell culture supernatant, passed through a 0.45-μm-pore-size filter, and stored at −80°C. To produce virus stocks from CEMx174 cells, the cultures were diluted with fresh medium and transfected with 3 μg of proviral DNA the following day, using a DEAE-dextran protocol (29). When cytopathic effects were observed, the cells were mixed with an equal volume of uninfected cells, pelleted, resuspended in fresh medium, and cultivated overnight. Thereafter, cells were pelleted again and the cell-free culture supernatant was filtered and stored in aliquots at −80°C. The concentration of p27 antigen was determined with a commercially available antigen capture kit (Innogenetics, Zwijndrecht, Belgium). The viral stocks were used to infect the human T-B hybrid cell line CEMx174, rPBMC, and herpesvirus saimiri-transformed T-cell lines.
Cell culture.
CEMx174 and COS-1 cells were maintained as described previously (13). Herpesvirus saimiri-transformed T-cell lines of human (CB15) and of rhesus macaque (MmHF7062A) origin were kept in a mixture of equal amounts of RPMI 1640 and GC medium (Vitromex, Vilshofen, Germany) supplemented with 10% FCS and 100 U of interleukin-2 per ml. rPBMC, obtained from healthy rhesus macaques that were seronegative for SIV, type D retroviruses, and simian foamy virus, were isolated by using lymphocyte separation medium (Organon Teknika Corporation, Durham, N.C.), stimulated for 2 to 3 days with 5 μg of phytohemagglutinin (PHA) per ml, and cultured in RPMI 1640 medium with 20% FCS and 50 U of interleukin-2 per ml.
Viral replication.
CEMx174, CB15, and MmHF7062A cells split 1:2 to 1:5 the previous day were infected with an aliquot of SIVmac containing 2 ng of viral p27 antigen. Activated rPBMC were infected with the same amount of SIVmac upon PHA stimulation as described above. Cell culture supernatants were sampled at regular intervals and stored at −80°C, and virus production was measured by reverse transcriptase assay as described elsewhere (31).
In vitro transcription and translation reactions.
Elf-1 and Ets-1 expression plasmids (39) were kindly provided by D. M. Markovitz and J. M. Leiden. The in vitro transcription and translation reactions were performed with a commercially available kit (Promega) as instructed by the manufacturer.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were carried out with double-stranded oligonucleotide probes. Complementary single-stranded oligonucleotides were custom made (Eurogentec, Seraing, Belgium), and double-stranded oligonucleotides were generated by annealing of complementary oligonucleotides. Prior to the annealing reaction, one oligonucleotide strand was [γ-32P]ATP end labeled by polynucleotide kinase (New England Biolabs, Schwalbach, Germany). Double-stranded oligonucleotides were purified by electrophoresis in 12% native polyacrylamide gels and eluted from the gel matrix in Tris-EDTA buffer (pH 8.0) overnight at 4°C. The following oligonucleotide probes were used (the sequences of the sense strands are shown in 5′→3′ orientation): HIV-2 PuB2 (CAGCTATACTTGGTCAGGGCAGGAAGTAACTA), murine sarcoma virus (MSV) (TGCGCGCTTCCGCTCTCCGAG), SIV PuB1 (TGTCAGAGGAAGAGGTTAG), SIV PuB2 (TGGCTGACAAGAAGGAAACTCGCTGAAACAGCA), mut-SF3 (TGGCTGACAAGAAGGGAGCTCGCTGAAACAGCA), mut-SF2 (TGGCTGACATATGGGAAACTCGCTGAAACAGCA), and mut-Elf (TGGCTGACAAGAGTTGAGCTCGCTGAAACAGCA (mutated positions are underlined). DNA-protein binding reactions using T-cell nuclear extract were carried out in 75 mM KCl–10 mM Tris (pH 7.5)–1 mM dithiothreitol (DTT)–1 mM EDTA–4% Ficoll and contained 20,000 cpm of labeled oligonucleotide probe, 10 μg of nuclear extract, 250 ng of double-stranded poly(dI-dC) (Pharmacia, Freiburg, Germany), and 250 ng of calf thymus DNA (Merck, Darmstadt, Germany) in a final volume of 20 μl. DNA-protein binding reactions using in vitro-translated Ets-1 and Elf-1 proteins were carried out in 75 mM KCl–10 mM Tris (pH 7.5)–1 mM DTT–1 mM EDTA–4% Ficoll and contained 20,000 cpm of labeled oligonucleotide probe, 6 μl of in vitro-translated protein, and 380 ng of double-stranded poly(dI-dC) in a final volume of 30 μl. For supershift analysis, the reaction mixture additionally contained 2 μl of Elf-1 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.). The reaction mixtures were incubated for 30 min at room temperature and subsequently separated in 4% nondenaturing polyacrylamide gels. The gels contained 0.25 mM KCl and 0.5 mM DTT and were run in 0.25× Tris-borate-EDTA buffer at 120 V for 4 h at 4°C. Immunoblots were probed with a 1:10,000 dilution of the Elf-1 antibody in conjunction with a 1:5,000 dilution of the commercially available anti-rabbit horseradish peroxidase-coupled antibody (Dianova, Hamburg, Germany). Immunoblots were developed by using an enhanced chemiluminescence detection system (Amersham, Chicago, Ill.) as specified by the manufacturer.
Nuclear extracts.
MmHF7062A cells (1 × 107 to 5 × 107) were lysed with 10 mM HEPES (pH 7.9)–1.5 mM MgCl2–10 mM KCl–0.5 mM DTT–0.5 mM phenylmethylsulfonyl fluoride (PMSF)–0.1% (vol/vol) Nonidet P-40. Nuclei were pelleted by centrifugation in a microcentrifuge (Biofuge 13; Heracus Sepratech) at 3,000 rpm for 15 min at 4°C and resuspended in 20 mM HEPES (pH 7.9)–0.42 M NaCl–1.5 mM MgCl2–0.2 mM EDTA–0.5 mM DTT–0.5 mM PMSF–8 μg of aprotinin per ml–2 μg of leupeptin per ml–25% (vol/vol) glycerol. Reaction mixtures were incubated for 15 min at 4°C. Nuclear debris was pelleted by centrifugation in a microcentrifuge (Biofuge 13) at 14,000 rpm for 15 min at 4°C, and the cleared supernatants were dialyzed for 4 h at 4°C against 20 mM HEPES (pH 7.9)–0.1 M KCl–0.2 mM EDTA–0.5 mM DTT–0.5 mM PMSF–20% (vol/vol) glycerol. Aliquots were stored at −80°C, and protein concentrations were assayed by using a commercial kit (Bio-Rad, Munich, Germany).
RESULTS
Transcriptional activities of mutated SIV LTRs.
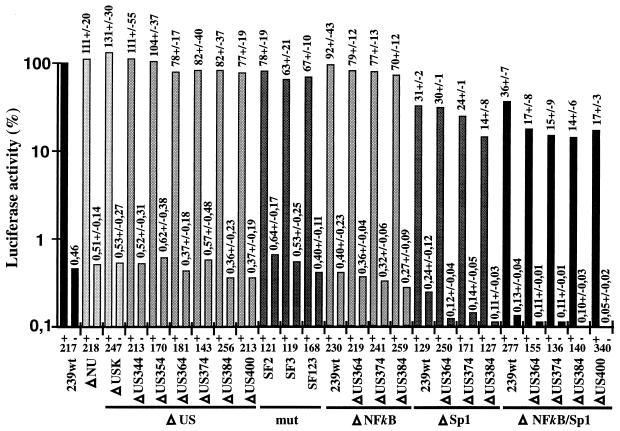
To investigate the influence of deletions and point mutations in the US region and the NF-κB and Sp1 binding sites on transcriptional activity, the luciferase reporter gene was expressed under the control of the mutated SIVmac239 LTRs in the presence or absence of Tat (Fig. 3). Removal of the 334 bp of US sequences that were selectively deleted in vivo had no significant effect (ΔNU, 111% ± 20% [Fig. 3]). Additional deletions of all U3 sequences upstream of the NF-κB binding site, except those important for integration, resulted in only very moderately decreased activity (ΔUS400, 77% ± 19% [Fig. 3]). Similar marginal effects were observed when the previously described factor binding sites SF1 to SF3 were mutated (mut-SF123, 67% ± 10% [Fig. 3]). In agreement with previously published data (14), deletion of the single NF-κB site had no significant effect (92% ± 43%), whereas deletion of all Sp1 binding sites resulted in an approximately threefold decrease of transcriptional activity in the presence of Tat (31% ± 2% [Fig. 3]). In the absence of the Sp1 sites, further deletion of 50 bp of the US65 region reduced reporter gene activity in the presence of Tat about two- to threefold, to approximately 15% of the 239wt LTR activity (ΔSp1/US384, 14% ± 8% [Fig. 3]). Even in the absence of the entire 400 bp of US sequences and all NF-κB and Sp1 binding sites, the truncated SIV promoter maintained approximately 15% of 239wt LTR activity and was responsive to Tat activation (ΔNF-κB/Sp1/US400, 17% ± 3% [Fig. 3]). Thus, under the experimental conditions used, deletion of about 87% of the SIVmac U3 region (452 bp), including the core enhancer and most of the US sequences, resulted in only a sixfold decrease of promoter activity and had little effect on Tat transactivation in COS-1 cells. Similar results were obtained after cotransfection of Jurkat T cells with a Tat expression vector and a luciferase reporter plasmid (ΔNU, 131% ± 34%; ΔUS400, 70% ± 43%; ΔNF-κB/US384, 79% ± 16%; ΔSp1/US384, 24% ± 13%; ΔNF-κB/Sp1, 46% ± 20%; ΔNF-κB/Sp1/US384, 19% ± 12%; numbers represent percentages of wild-type reporter activity obtained from six transfections). The absolute values were lower and more variable, however, than those obtained for COS-1 cells.
FIG. 3.
Activities of mutated SIVmac239 promoters with (+) or without (−) addition of a Tat expression plasmid. COS-1 cells were transfected with the indicated LTR-luciferase plasmids alone or cotransfected with an SIVmac Tat expression plasmid. For better comparison between independent experiments, stock DNA preparations from the wild-type LTR-luciferase and SIVmac Tat expression plasmids were always included in duplicate as positive controls (100% activity). Promoter activities relative to the 239wt promoter together with values for standard errors are shown above the bars. The results were obtained from four to six independent experiments. Background values for the luciferase plasmid without SIV LTR sequences were 0.16% ± 0.06% in the presence and 0.1% ± 0.04% in the absence of Tat. Background values were deducted from the measurements. Numbers at the bottom indicate the ratios of luciferase activities observed in the presence and in the absence of Tat. The ΔUSK mutant contains a 334-bp deletion in the US region, which is also present in ΔNU, and SstII and XhoI sites just upstream of the US65 region.
Virus production in COS-1 cells.
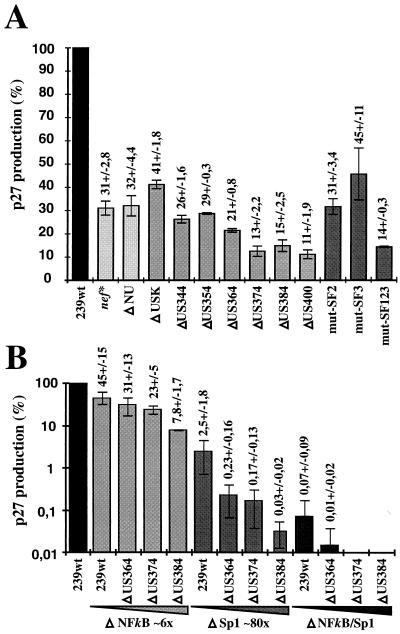
Full-length proviral SIVmac239 constructs, containing mutations in both LTRs to prevent recombination, were transfected into COS-1 cells for virus production. A defective nef gene resulted in an approximately threefold reduction of p27 core antigen production compared to nef-open SIVmac239 (nef*, 31% ± 2.8% [Fig. 4A]). The deletion of 354 bp of upstream U3 sequences had no significant effect (ΔUS354, 29% ± 0.3% [Fig. 4A]). Further deletion of all U3 sequences upstream of the NF-κB site, except those required for integration, reduced p27 production about threefold compared to SIVmac239 nef* (ΔUS400, 11% ± 1.9% [Fig. 4A]). Mutating the previously described SF1 to SF3 binding sites in the SIVmac239 ΔNU variant resulted in an approximately twofold decrease of p27 production compared to nef-defective SIVmac239 (mut-SF123, 14% ± 0.3% [Fig. 4A]). Thus, deletion and mutation of the SF1 to SF3 sites had comparable effects on p27 production (ΔUS384, 15% ± 2.5%; mut-SF123, 14% ± 0.3% [Fig. 4A]).
FIG. 4.
Production of p27 core antigen by COS-1 cells transfected with proviral constructs containing mutations in both LTRs. (A) p27 production measured after transfection with clones containing deletions in the US65 region, mutations in the SF binding sites, or a premature in-frame TAA stop signal at codon 93 of nef (nef*) (19). (B) Virus production obtained after transfection with proviral constructs bearing upstream U3 deletions in conjunction with NF-κB and/or Sp1 deletions. For clarity, panel B is shown in a logarithmic scale. Stock preparations of 239wt DNA were always transfected in parallel, and virus production is shown as a percentage of the wild-type value. Exact values with standard deviations are indicated above the bars. Average p27 production obtained with wild-type virus was about 50 ng of p27 per ml. The results are mean values obtained from four independent experiments.
In the presence of the NF-κB and Sp1 binding sites, deletion of the 60 bp just upstream of the core enhancer reduced virus production about threefold (Fig. 4A). To assess the relevance of the US region for virus production in the absence of core enhancer elements, SIVmac239 LTR variants containing changes in both the US region and the core enhancer elements were also investigated (Fig. 4B). Deletion of the Sp1 sites reduced p27 production about 40-fold (2.5% ± 1.8%), whereas removal of the single NF-κB site had only a 2-fold effect (45% ± 15%) (Fig. 4B). In the absence of the NF-κB site, an additional deletion of 374 bp of US sequences had little effect, and virus production was comparable to that of nef-defective SIVmac239 (ΔNF-κB/US374, 23% ± 5%, and nef*, 31% ± 2.8% of the value for nef-open SIVmac [Fig. 4]). In contrast, deletion of these US sequences resulted in approximately 15-fold-lower p27 levels in the absence of the four Sp1 sites (ΔSp1, 2.5% ± 1.8%; ΔSp1/US374, 0.17% ± 0.13%). Deletion of the region encompassing the SF2 and SF3 binding sites (US384) had a four- to sixfold negative effect on p27 antigen production in the absence of NF-κB or Sp1 binding sites (Fig. 4B). When the entire core enhancer was deleted, virus production was reduced more than 1,000-fold (ΔNF-κB/Sp1, 0.07% ± 0.09% compared to 239wt). The amount of p27 obtained after transfection of COS-1 with the ΔNF-κB/Sp1 mutant was close to the detection limit of approximately 20 pg of p27 antigen per ml, and no significant virus production could be detected when additional upstream sequences were deleted (ΔNF-κB/Sp1/US364, 0.01% compared to 239wt [Fig. 4B]). Thus, with the same cell line and transfection protocol, the effect of deletions of the core enhancer region on virus production was more than 2 orders of magnitude higher than the effect on transcriptional activity in transient reporter assays (Fig. 3 and 4B). The deletion of 384 bp of the US region had little effect on luciferase reporter gene expression in the presence or absence of the core enhancer elements (Fig. 3). Nonetheless, this deletion resulted in a sixfold decrease in virus production in the context of an NF-κB-deleted provirus (ΔNF-κB, 45% ± 15%; ΔNF-κB/US384, 7.8% ± 1.7%). An 80-fold drop in p27 antigen levels was observed when the four Sp1 binding sites were removed (ΔSp1, 2.5% ± 1.8%; ΔSp1/US384, 0.03% ± 0.02% [Fig. 4B]).
Replication of SIVmac239 LTR mutants.
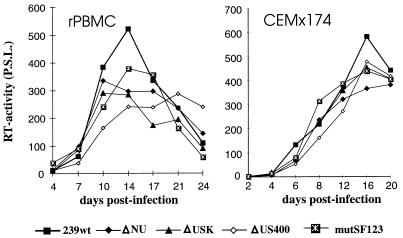
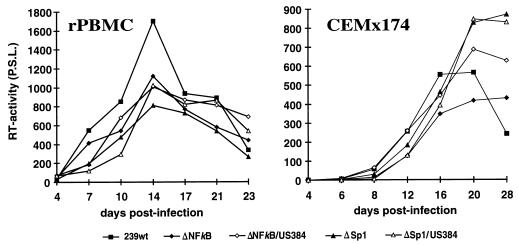
Virus stocks prepared from transient transfection of COS-1 cells were used to investigate the replicative properties of the SIVmac239 variants with deletions in the US region or mutations in the SF1 to SF3 factor binding sites. All mutants replicated with kinetics highly similar to those of the parental SIVmac239 clone in PHA-stimulated rPBMC, CEMx174 cells, MmHF7062A cells, and CB15 cells (Fig. 5 and data not shown). Thus, deletion of essentially all U3 sequences upstream of the NF-κB sites (400 bp) had no significant effect on replication of SIVmac. In the next set of experiments, we analyzed the replicative properties of SIVmac239 LTR mutants containing deletions in the US region in conjunction with deletions of the NF-κB or Sp1 binding elements. COS-1 transfection with proviral constructs missing the Sp1 binding sites resulted in inefficient virus production (Fig. 4B). Therefore, virus stocks generated from transfected CEMx174 cells were used for these experiments. PCR and sequence analysis at the end of culture confirmed the presence of the deletions (data not shown). All US deletion mutants replicated with efficiencies similar to that for the parental SIVmac239 clone in the four cell lines mentioned above when the single NF-κB binding site or the Sp1 binding sites were preserved (Fig. 6 and data not shown). The effective replication of the LTR variants containing US deletions in conjunction with the Sp1 deletion was striking, because these LTR variants showed up to 3,000-fold-lower amounts of p27 antigen production in transfected COS-1 cells (ΔSp1/US384, 0.03% ± 0.02% [Fig. 4B and 6]).
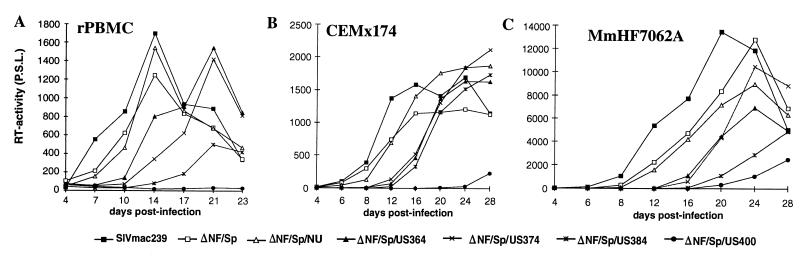
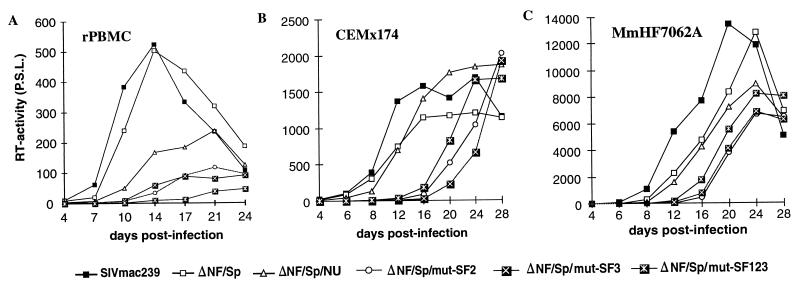
FIG. 5.
Replication of SIVmac239 US and SF mutants in rPBMC and CEMx174 cells. Virus containing 2 ng of p27 derived from transfected COS-1 cells was used for infection. Similar results were obtained in three independent experiments. RT, reverse transcriptase; P.S.L., photo-stimulated luminescence.
FIG. 6.
Replication of SIVmac239 LTR variants containing deletions in the US region in conjunction with NF-κB or Sp1 site deletions. Virus containing 2 ng of p27 derived from transfected CEMx174 cells was used for infection. The results were confirmed in four independent experiments. For abbreviations, see the legend to Fig. 5.
Next, we investigated if enhancer elements located in the US65 region have an effect on SIVmac replication when the entire core enhancer region is deleted. In agreement with previously published data (14), the NF-κB/Sp1 deletion had little effect on replication (ΔNF/Sp [Fig. 7]). Also, deletion of the 334 bp of upstream U3 sequences that were not preserved in vivo (22), in conjunction with the core enhancer region, did not reduce the replicative efficiency (ΔNF/Sp/NU [Fig. 7]). When the first 30 to 40 bp of the US65 region, including the SF1 site, were also removed, delayed replication kinetics were observed in rPBMC, CEMx174 cells, and MmHF7062A cells (ΔNF/Sp/US364 and ΔNF/Sp/US374 [Fig. 7]). Additional deletion of the next 10 bp (ΔNF/Sp/US384), affecting the SF2 and SF3 binding sites, further delayed and reduced virus production in rPBMC and MmHF7062A cells (Fig. 7A and C). In contrast, the ΔNF/Sp/US384 variant replicated with kinetics highly similar to those of the ΔNF/Sp/US364 and ΔNF/Sp/US374 variants in CEMx174 cells (Fig. 7B). The ΔNF/Sp/US400 variant, in which only 65 bp of the 517 bp U3 region (13%) are preserved, showed no detectable levels of reverse transcriptase activity in rPBMC (Fig. 7A). Low levels of replication, however, could be measured after infection of CEMx174 and MmHF7062A cells (Fig. 7B and C). Thus, SIVmac239 with only 95 of the 517 bp of the SIVmac U3 region (18%), containing only sequences required for integration, the TATA box, and a single NF-κB binding site, grows with normal kinetics in the four cell types analyzed (ΔSp1/US384 [Fig. 6 and data not shown]). Only the presence of the 26 bp usually located immediately upstream of the NF-κB site was required for efficient viral replication in the absence of the entire core enhancer (Fig. 7). The results show that enhancer elements located in the 26 bp upstream of the NF-κB site can compensate for the complete loss of the core enhancer.
FIG. 7.
Replication of SIVmac239 LTR variants containing deletions in the US region in conjunction with a deletion of all NF-κB and Sp1 sites. Virus containing 2 ng of p27 derived from transfected CEMx174 cells was used for infection of rPBMC (A), CEMx174 (B), and MmHF7062A (C) cells. For abbreviations, see the legend to Fig. 5.
Deletion of the SF2 and SF3 binding sites reduced viral replication in rPBMC and MmHF7062A cells (Fig. 7). Therefore, we tested if point mutations in these factor binding sites also affect replication. As shown in Fig. 8, mutations altering the SF2 (AGAAGG→TATGGG) or SF3 (GGAAA→ GG G AG) binding site resulted in delayed replication in CEMx174 and MmHF7062A cells compared to the ΔNF/Sp/NU control virus (Fig. 8). In these two cell lines, the variant containing mutations in all three SF sites replicated with an efficiency comparable to those of the ΔNF/Sp/mut-SF2 and ΔNF/Sp/mut-SF3 mutants. Mutations in the SF2 or SF3 site also reduced replication in rPBMC (Fig. 8A). The effects varied slightly between experiments performed with rPBMC from different animals. Nonetheless, these point mutations always caused a delay in viral replication, with the most drastic effect occurring when all three sites were altered.
FIG. 8.
Replication of SIVmac239 LTR variants containing point mutations in the SF1 to SF3 binding sites in conjunction with a deletion of all NF-κB and Sp1 sites. Virus containing 2 ng of p27 derived from transfected CEMx174 cells was used for infection of rPBMC (A), CEMx174 (B), and MmHF7062A (C) cells. In about one-third of the experiments we observed a less efficient replication of nef-defective SIVmac239 even in PHA-stimulated rPBMC. Replication of the ΔNF/Sp/NU variant was always comparable to that of the nef* variant, indicating that the slightly reduced and delayed replication kinetics are a Nef and not an LTR effect. For abbreviations, see the legend to Fig. 5.
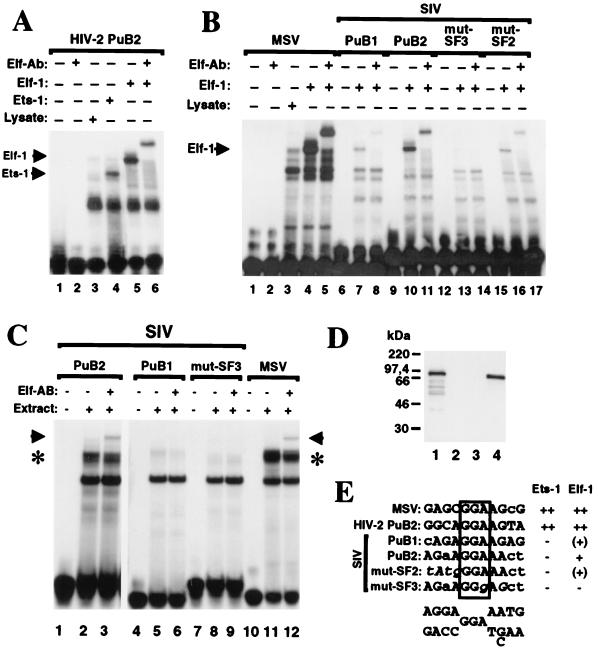
The PuB2 site binds to Elf-1.
It has previously been demonstrated that two PuB sites located in the HIV-2 enhancer bind to Elf-1 and Ets-1 (24–26). SIVmac is closely related to HIV-2, and two purine-rich sequences can be identified at a similar location in the SIV enhancer (Fig. 2). One of these purine-rich regions, named SIV PuB1 in analogy to the HIV-2 LTR, is located just at the 3′ end of the NOD and was not conserved in the majority of animals infected with nef-defective SIVmac239 (reference 22 and Fig. 2). EMSAs with in vitro-translated Ets-1 and Elf-1 or cellular nuclear extracts were performed to test binding to SIV PuB1 and PuB2 (Fig. 9). Although the SIV PuB1 site is highly similar to the consensus binding site for members of the Ets family of transcription factors (Fig. 9E), it bound only weakly to Elf-1 (Fig. 9B, lane 7) and not to Ets-1. The presence or absence of the PuB1 site had no effect on viral replication or promoter activity (data not shown). The SIVmac PuB2 site completely overlaps the previously defined SF2 and SF3 binding sites (40) (Fig. 2 and 9E). It binds to Elf-1 (Fig. 9B, lane 10) but, in contrast to the HIV-2 PuB2 site, not to Ets-1 (Fig. 9A, lane 4, and data not shown). Binding to Elf-1 was reduced compared to the MSV and HIV- 2 PuB control probes (Fig. 9A, lane 5; Fig. 9B, lanes 4 and 10). Mutations in the conserved central GGA motif (SF3) abolished, and changes in the 5′ flanking region (SF2) of the SIV PuB2 site reduced, Elf-1 binding (Fig. 9B, lanes 13 and 16). Supershift assays with Elf-1 antibody confirmed that the PuB2 binding protein was indeed Elf-1 (Fig. 9B, lane 11). To demonstrate that Elf-1 binds also in the presence of other DNA binding factors, further gel retardation experiments were performed with nuclear extracts from MmHF7062A cells (Fig. 9C). As shown in Fig. 9C, lanes 2 and 11, incubation of nuclear extracts with probes corresponding to the PuB2 and the MSV probe resulted in similar band shifting patterns. While the lower band could also be observed with the PuB1 and mut-SF3 probe, the upper complex was specific for the MSV and PuB2 probes (Fig. 9C, lanes 2, 5, 8, and 11, upper complex indicated by asterisk). Addition of a monoclonal antibody which reacted specifically with Elf-1 (Fig. 9D) gave rise to a supershifted complex with the PuB2 and MSV probes but not with the PuB1 or mutSF3 oligonucleotide (Fig. 9C, lanes 3, 6, 9, and 12; supershift indicated by arrow). However, even in the presence of high amounts of Elf-1 antibody, the PuB2- and MSV-specific complex could not be disrupted entirely, indicating the binding of additional nuclear proteins to these sites (data not shown). We conclude from these data that Elf-1 binding to the PuB2 site can even be detected in the presence of a complex mixture of DNA binding proteins, but further, as yet unidentified factors contribute to the formation of nucleoprotein complexes on this sequence element.
FIG. 9.
Binding of Elf-1 to the SIVmac239 PuB sites. (A) Elf-1 and Ets-1 bind to the HIV-2 PuB2 element. In vitro-transcribed and -translated Ets-1 (lane 4) and Elf-1 (lane 5), and as a control reticulocyte lysate (lane 3), were incubated with a radiolabeled probe corresponding to the HIV-2 PuB2 site (see Materials and Methods). The Elf-1 extract was incubated with an antibody directed against Elf-1 (Elf-Ab; lane 6). (B) Elf-1 binding to SIV PuB sites. In vitro-transcribed and -translated Elf-1 and a control reticulocyte lysate were incubated with the radiolabeled MSV and SIV probes in the presence or absence of Elf-1 antibody as indicated. The specific probes are described in Materials and Methods. (C) Elf-1 binds to the SIV PuB2 site in MmHF7062A cells. Extracts from MmHF7062A cells were incubated with the SIV PuB2 probe in the presence or absence of Elf-specific antibody as indicated. Asterisks indicate PuB2- and MSV-specific complexes. Arrows indicate supershift by Elf-1 antibody. (D) The Elf-1 antibody is highly specific. In vitro-transcribed and -translated Elf-1 (lane 1), a control reticulocyte lysate (lane 2), in vitro-transcribed and -translated Ets-1 (lane 3), and nuclear extracts from MmHF7062A cells were separated on a sodium dodecyl sulfate–12.5% polyacrylamide gel, immunoblotted, and probed with Elf-1 antibody as described in Materials and Methods. (E) Purine-rich sequences in the probes used for EMSA. The conserved central GGA motif is boxed; the consensus binding site for members of the Ets family of transcription factors (39) is indicated at the bottom. Results of the EMSA are shown at the right [++, strong binding; +, binding; (+), weak binding; −, no binding].
DISCUSSION
In contrast to HIV-1, SIVmac without all Sp1 and NF-κB elements replicates efficiently in lymphoid culture and is even capable of causing AIDS in rhesus macaques (14, 15). Furthermore, 334 bp of upstream LTR U3 sequences seem not to be advantageous in vivo, since they are selectively deleted after infection with nef-defective SIVmac239 (22). These findings suggested the presence of important enhancer elements in the remaining 65 bp just upstream of the SIVmac LTR core enhancer elements. Therefore, it was unexpected that deletion of the entire upstream U3 region of 400 bp had very little effect on promoter activity and on viral replication in lymphoid cells. Even mutants missing 384 bp of the upstream region and the Sp1 or NF-κB sites replicated with kinetics and to levels similar to those of 239wt in various cell lines and rPBMC cultures.
It has been previously suggested that the sequences located immediately upstream of the NF-κB element account for the high activity of SIVmac239 missing the entire core enhancer region (14, 15). In the absence of all Sp1 elements and the NF-κB binding site, we could clearly demonstrate and partly characterize this novel upstream regulatory element. Deletion of up to 364 bp of US sequences had little effect on viral replication, even in the absence of the entire core enhancer (Fig. 10). Deletions or point mutations affecting the remaining 26 bp of the US region containing the previously described SF2 and SF3 sites (40) clearly delayed and reduced viral replication in herpesvirus saimiri-transformed T-cell lines and rPBMC. The purine-rich sequences encompassing the SF2 and SF3 sites show high homology to the DNA recognition sites of members of the Ets family of transcription factors (39). We could show that the SIVmac239 PuB2 element binds efficiently to Elf-1, a lymphoid-specific Ets transcription factor that regulates inducible gene expression during T-cell activation (39). In contrast to the PuB2 region of the closely related HIV-2, however, no binding to Ets-1, another member of the Ets family of proto-oncogene products, could be observed. The specificity for Elf-1 binding is not unexpected, since it has been previously demonstrated that an A rather than T at position 8 of the binding site mediates Elf-1 binding (39). It is worth noting that the enhancer of HIV-2 contains two PuB sites that bind to Elf-1 and/or Ets-1 (24, 26) and that a second purine-rich region that resembles an Ets binding site is also present in the SIVmac239 enhancer. The upstream SIVmac239 PuB1 site was affected by the upstream U3 deletions in six of nine monkeys experimentally infected with nef-defective SIVmac239 (22). Interestingly, however, these deletions always created new putative purine boxes (underlined) (Δ9650-9800 [ATGAGGAGΔAAG] and Δ9521-9809 [AAAAGΔGAAGAA]; numbers refer to the SIVmac239 sequence published by Regier and Desrosiers [32]; Δ indicates the position of the deletion selected in vivo). Moreover, these Ets binding motifs are conserved among most SIV LTR sequences in the database (27). The exceptions are two unusual molecular clones, the SIVmac142 clone, which is not infectious for macaques (29), and the acutely pathogenic SIVPBj14 clone (5, 8). The results suggest that the presence of a second purine-rich site may be advantageous in vivo. In cell culture, however, deletion of the PuB1 site had little or no effect on replication.
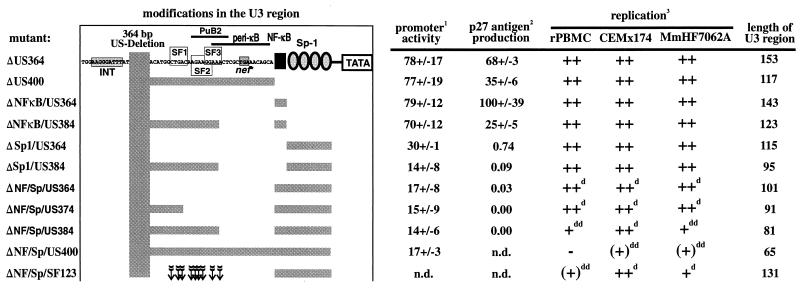
FIG. 10.
Biological effects of some mutations in the SIVmac239 U3 region investigated in this study. 1Promoter activity in the presence of Tat in COS-1 cells. 2p27 antigen production in COS-1 cells relative to the nef-defective SIVmac239 nef* variant. 3−, no replication detected; (+) maximal levels of virus production detected were <10% of the parental 239wt clone; +, maximal levels of virus production detected were 10 to 50% of the original clone; ++, virus production comparable to that of 239wt; d, delayed growth kinetics; dd, strongly delayed growth kinetics. The arrows indicate the positions of points mutations in the SF123 mutant.
Some work has been done to define and characterize the SIVmac and HIV-2 enhancers (24–26, 33, 39, 40). Most of these studies, however, have focused exclusively on the characterization of factor binding sites or the investigation of transcriptional activity in transient assays. In the present study, the effects of deletions and mutations on transcriptional activity, virus production, replication, and factor binding of the SIVmac239 LTR were analyzed in parallel. The impact of LTR mutations on virus production was much higher than the effects on reporter gene expression in transiently transfected COS-1 cells. Virus production is obviously a more complex process than reporter gene expression and depends on cooperative effects of several regulatory factors. Perhaps less efficient activation and transport of unspliced RNA due to about sixfold-reduced expression of both Tat and Rev may explain the drastically reduced levels of virus production. Deletion of the US region and the Sp1 sites reduced p27 production in COS-1 cells more than 1,000-fold without having a significant effect on replication in CEMx174 cells and rPBMC. Thus, the high levels of activated NF-κB proteins in these cell types seem to be sufficient to allow efficient viral replication.
It seems that as for HIV-2 (24–26), Elf-1 binding may be important for transactivation of SIVmac gene expression in lymphoid cells. Some differences, however, should also be noted. Binding of SIV PuB1 and PuB2 to Elf-1 was relatively weak compared to the HIV-2 and MSV controls. It has been suggested, however, that such low-affinity Elf-1 sites may be of particular importance in the regulation of lymphoid-specific gene expression (16). A TG-rich element, named the pets site, has been identified in the HIV-2 enhancer (7) but is absent in the SIVmac239 LTR. Interestingly, even when 384 bp of US sequences, including the two purine-rich Elf-1 binding sites and the core enhancer elements, were removed, significant levels of viral replication were observed, particularly in CEMx174 cells (Fig. 10). Deletion of the remaining 16 bp of US sequences, however, strongly impaired viral replication, indicating that another enhancer element is located in this region. This sequence shows substantial sequence similarity to the corresponding region in the HIV-2 LTR, designated the peri-κB site, which binds to nuclear factors from PBMC and T cells (2). In transient transactivation assays, activation of the HIV-2 enhancer by the peri-κB site was only observed in monocytes and not in T cells (2). Our results obtained with rPBMC and herpesvirus saimiri-transformed T-cell lines of both human and rhesus origin indicate that this region increases the promoter activity of SIVmac239 in T cells.
It was very surprising that highly conserved enhancer elements, like the Sp1 and NF-κB binding sites, are largely dispensable for the pathogenicity of primate lentiviruses. It seems that the functional organization of the SIVmac enhancer differs considerably from that of the HIV-1 enhancer. The relative importance of the Elf-1 binding and the peri-κB sites for replication in differential cell types, and most importantly for viral pathogenicity, remains to be elucidated.
ACKNOWLEDGMENTS
We thank David M. Markovitz and Jeffrey M. Leiden for reagents and helpful comments, Marion Hamacher for excellent technical assistance, Helmut Fickenscher for herpesvirus saimiri-transformed T-cell lines, Bernhard Fleckenstein and Ronald C. Desrosiers for support, and Klaus Überla for helpful suggestions.
This work was supported by SFB 466, SFB 473, grant Sta 357/3-1/SFB473, and a fellowship from the German BMBF to F.K.
REFERENCES
- 1.Bassuk A G, Anandappa R T, Leiden J M. Physical interactions between Ets and NF-κB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark N M, Hannibal M C, Markovitz D M. The peri-κB site mediates human immunodeficiency virus type 2 enhancer activation in monocytes but not in T cells. J Virol. 1995;69:4854–4862. doi: 10.1128/jvi.69.8.4854-4862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–703. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 4.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 5.Dewhurst S, Embretson J E, Anderson D C, Mullins J I, Fultz P N. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature. 1990;345:636–640. doi: 10.1038/345636a0. [DOI] [PubMed] [Google Scholar]
- 6.di Fagagna F D, Marzio G, Gutierrez M I, Kang L Y, Falaschi A, Giacca M. Molecular and functional interactions of transcription factor USF with the long terminal repeat of human immunodeficiency virus type 1. J Virol. 1995;69:2765–2775. doi: 10.1128/jvi.69.5.2765-2775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu G K, Markovitz D M. Purification of the pets factor. A nuclear protein that binds to the inducible TG-rich element of the human immunodeficiency virus type 2 enhancer. J Biol Chem. 1996;271:19599–19605. doi: 10.1074/jbc.271.32.19599. [DOI] [PubMed] [Google Scholar]
- 8.Fultz P N, McClure H M, Anderson D C, Switzer W M. Identification and biologic characterization of an acutely lethal variant of simian immunodeficiency virus from sooty mangabeys (SIV/SMM) AIDS Res Hum Retroviruses. 1989;5:397–409. doi: 10.1089/aid.1989.5.397. [DOI] [PubMed] [Google Scholar]
- 9.Garcia, J. A., D. Harrich, E. Soultanakis, F. Wu, R. Mitsuyasu, and R. B. Gaynor. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 8: 765–778. [DOI] [PMC free article] [PubMed]
- 10.Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992;6:347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Gundlach B R, Linhart H, Dittmer U, Sopper S, Reiprich S, Fuchs D, Fleckenstein B, Hunsmann G, Stahl-Hennig C, Überla K. Construction, replication, and immunogenic properties of a simian immunodeficiency virus expressing interleukin 2. J Virol. 1997;71:2225–2232. doi: 10.1128/jvi.71.3.2225-2232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrich D, Garcia J, Wu F, Mitsuyasu R, Gonazalez J, Gaynor R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1989;63:2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilyinskii P O, Daniel M D, Simon M A, Lackner A A, Desrosiers R C. The role of upstream U3 sequences in the pathogenesis of simian immunodeficiency virus-induced AIDS in rhesus monkeys. J Virol. 1994;68:5933–5944. doi: 10.1128/jvi.68.9.5933-5944.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilyinskii P O, Desrosiers R C. Efficient transcription and replication of simian immunodeficiency virus in the absence of NF-κB and Sp1 binding elements. J Virol. 1996;70:3118–3126. doi: 10.1128/jvi.70.5.3118-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilyinskii P O, Simon M A, Czajak S C, Lackner A A, Desrosiers R C. Induction of AIDS by simian immunodeficiency virus lacking NF-κB and Sp1 binding elements. J Virol. 1997;71:1880–1887. doi: 10.1128/jvi.71.3.1880-1887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.John S, Marais R, Child R, Light Y, Leonard W J. Importance of low affinity Elf-1 sites in the regulation of lymphoid-specific inducible gene expression. J Exp Med. 1996;183:743–750. doi: 10.1084/jem.183.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones K A, Kadonaga J T, Luciw P A, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 18.Kawakami K, Scheidereit C, Roeder R G. Identification and purification of a human immunoglobulin-enhancer-binding protein (NF-kappa B) that activates transcription from a human immunodeficiency virus type 1 promoter in vitro. Proc Natl Acad Sci USA. 1988;85:4700–4704. doi: 10.1073/pnas.85.13.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 20.Kharroubi A E, Martin M A. cis-acting sequences located downstream of the human immunodeficiency virus type 1 promoter affect its chromatin structure and transcriptional activity. Mol Cell Biol. 1996;16:2958–2966. doi: 10.1128/mcb.16.6.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J Y, Gonzalez-Scarano F, Zeichner S L, Alwine J C. Replication of type 1 human immunodeficiency viruses containing linker substitution mutations in the −201 to −130 region of the long terminal repeat. J Virol. 1993;67:1658–1662. doi: 10.1128/jvi.67.3.1658-1662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchhoff F, Kestler III H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 24.Leiden J M, Wang C Y, Petryniak B, Markovitz D M, Nabel G J, Thompson C B. A novel Ets-related transcription factor, Elf-1, binds to human immunodeficiency virus type 2 regulatory elements that are required for inducible trans activation in T cells. J Virol. 1992;66:5890–5897. doi: 10.1128/jvi.66.10.5890-5897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markovitz D M, Hannibal M, Perez V L, Gauntt C, Folks T M, Nabel G J. Differential regulation of human immunodeficiency viruses (HIVs): a specific regulatory element in HIV-2 responds to stimulation of the T-cell antigen receptor. Proc Natl Acad Sci USA. 1990;87:9098–9102. doi: 10.1073/pnas.87.23.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markovitz D M, Smith M J, Hilfinger J, Hannibal M C, Petryniak B, Nabel G J. Activation of the human immunodeficiency virus type 2 enhancer is dependent on purine box and kappa B regulatory elements. J Virol. 1992;66:5479–5484. doi: 10.1128/jvi.66.9.5479-5484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers G, Korber B, Berzofsky J A, Smith R F, Pavlakis G N. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1993. [Google Scholar]
- 28.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 29.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pazin M J, Sheridan P L, Cannon K, Cao Z, Keck J G, Kadonaga J T, Jones K A. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 31.Potts B J. “Mini” reverse transcriptase (RT) assay. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton; 1990. pp. 103–106. [Google Scholar]
- 32.Regier D A, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 33.Renjifo B, Speck N A, Winandy S, Hopkins N, Li Y. cis-acting elements in the U3 region of a simian immunodeficiency virus. J Virol. 1990;64:3130–3134. doi: 10.1128/jvi.64.6.3130-3134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross E K, Buckler-White A J, Rabson A B, Englund G, Martin M A. Contribution of NF-κB and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J Virol. 1991;65:4350–4358. doi: 10.1128/jvi.65.8.4350-4358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 36.Suñè C, García-Blanco M A. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:6572–6576. doi: 10.1128/jvi.69.10.6572-6576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor-α enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 38.Varmus H. Retroviruses. Science. 1988;240:1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- 39.Wang C Y, Petryniak B, Ho I C, Thompson C B, Leiden J M. Evolutionarily conserved Ets family members display distinct DNA binding specificities. J Exp Med. 1993;175:1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winandy S, Renjifo B, Li Y, Hopkins N. Nuclear factors that bind two regions important for transcriptional activity of the simian immunodeficiency virus long terminal repeat. J Virol. 1992;66:5216–5223. doi: 10.1128/jvi.66.9.5216-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeichner S L, Kim J Y H, Alwine J C. Linker-scanning mutational analysis of the transcriptional activity of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1991;65:2436–2444. doi: 10.1128/jvi.65.5.2436-2444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]