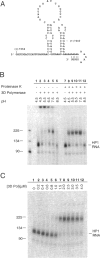
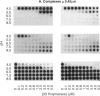
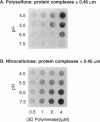
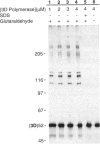
Abstract
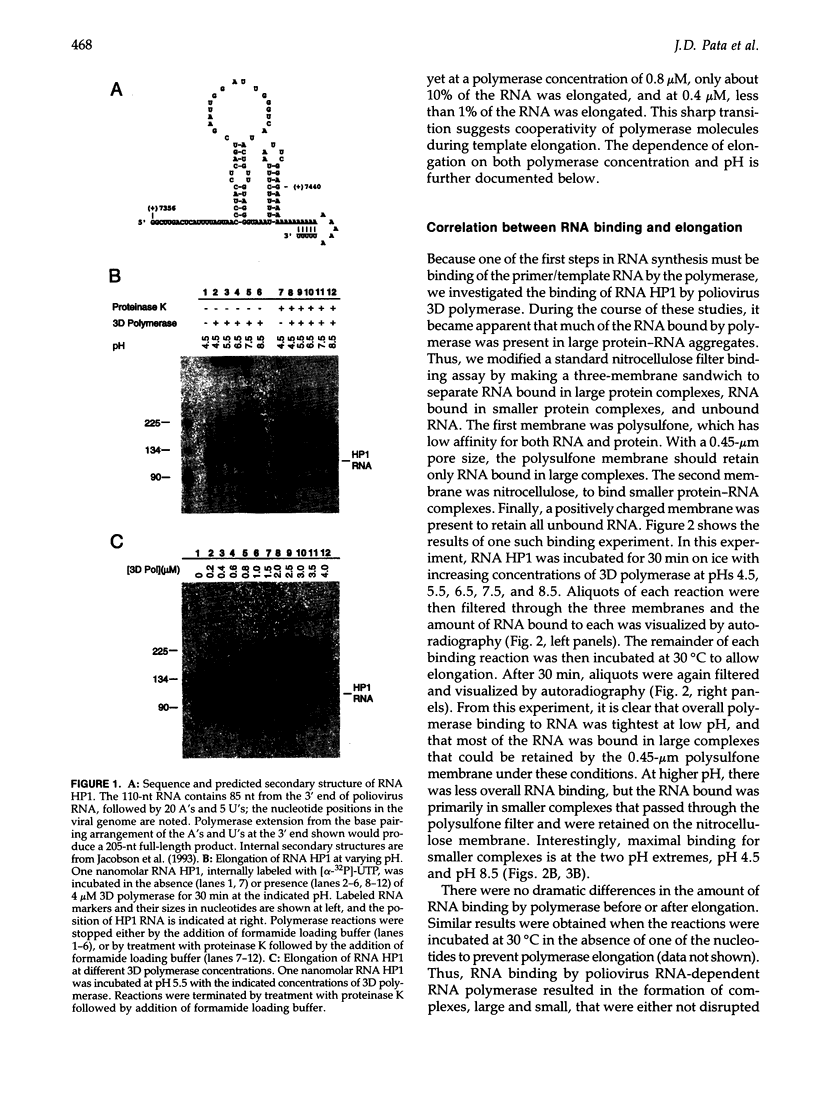
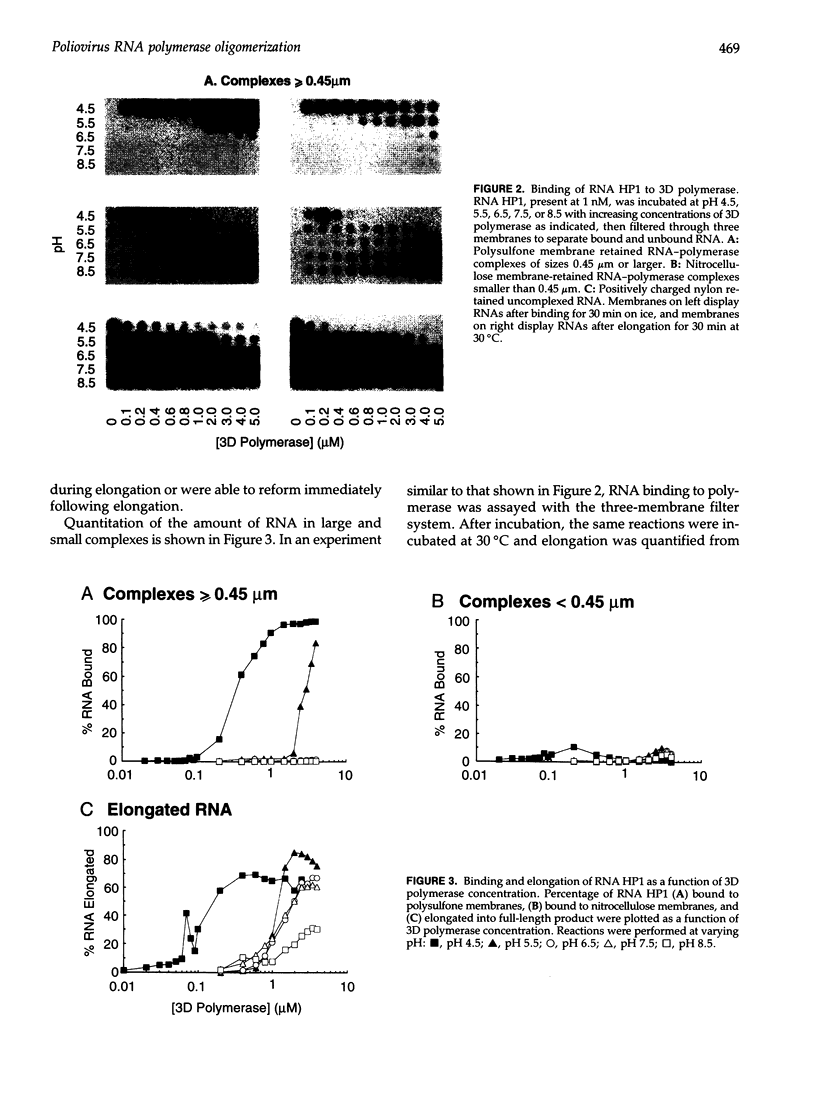
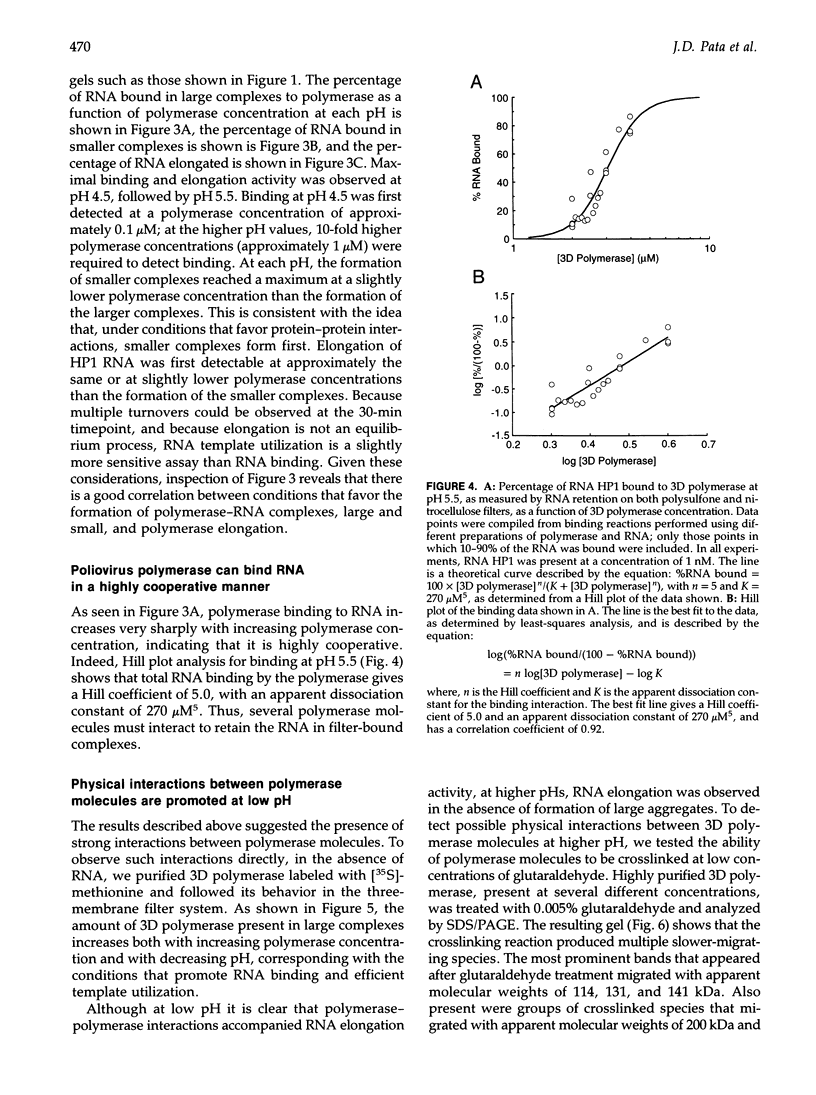
Using a hairpin primer/template RNA derived from sequences present at the 3' end of the poliovirus genome, we investigated the RNA-binding and elongation activities of highly purified poliovirus 3D polymerase. We found that surprisingly high polymerase concentrations were required for efficient template utilization. Binding of template RNAs appeared to be the primary determinant of efficient utilization because binding and elongation activities correlated closely. Using a three-filter binding assay, polymerase binding to RNA was found to be highly cooperative with respect to polymerase concentration. At pH 5.5, where binding was most cooperative, a Hill coefficient of 5 was obtained, indicating that several polymerase molecules interact to retain the 110-nt RNA in a filter-bound complex. Chemical crosslinking with glutaraldehyde demonstrated physical polymerase-polymerase interactions, supporting the cooperative binding data. We propose a model in which poliovirus 3D polymerase functions both as a catalytic polymerase and as a cooperative single-stranded RNA-binding protein during RNA-dependent RNA synthesis.
Full text
PDF





Images in this article
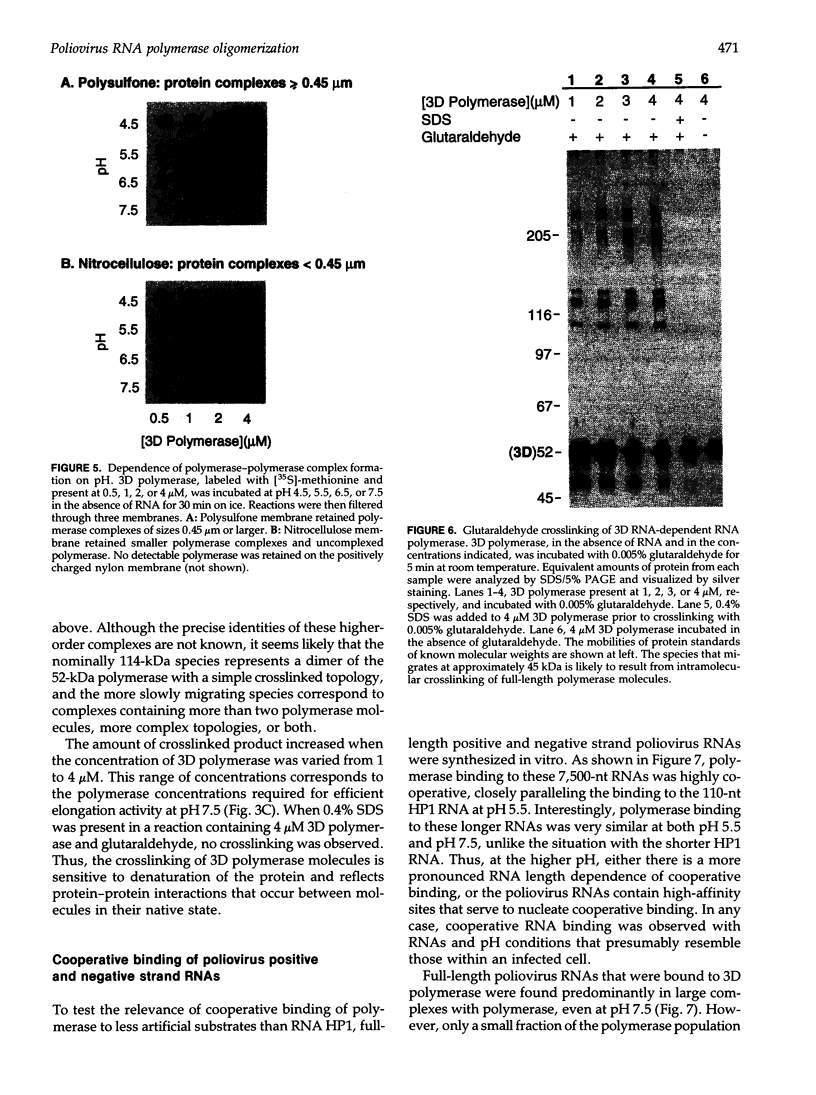
Selected References
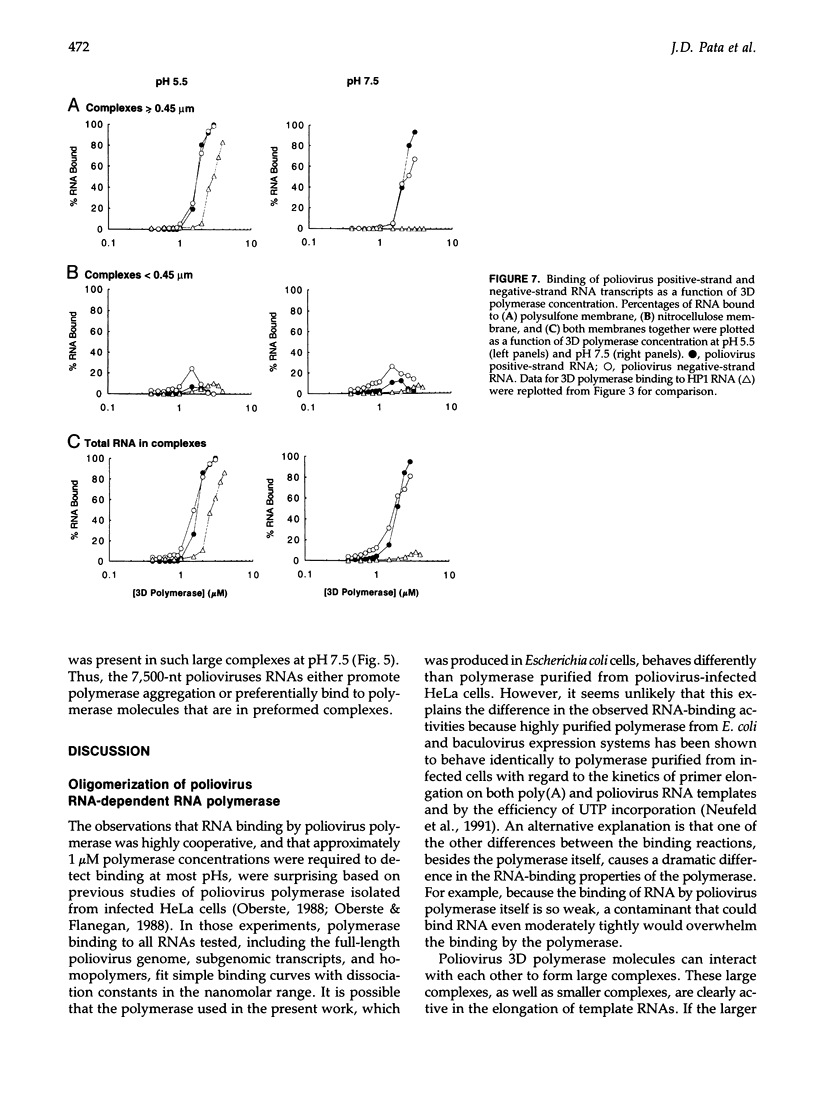
These references are in PubMed. This may not be the complete list of references from this article.
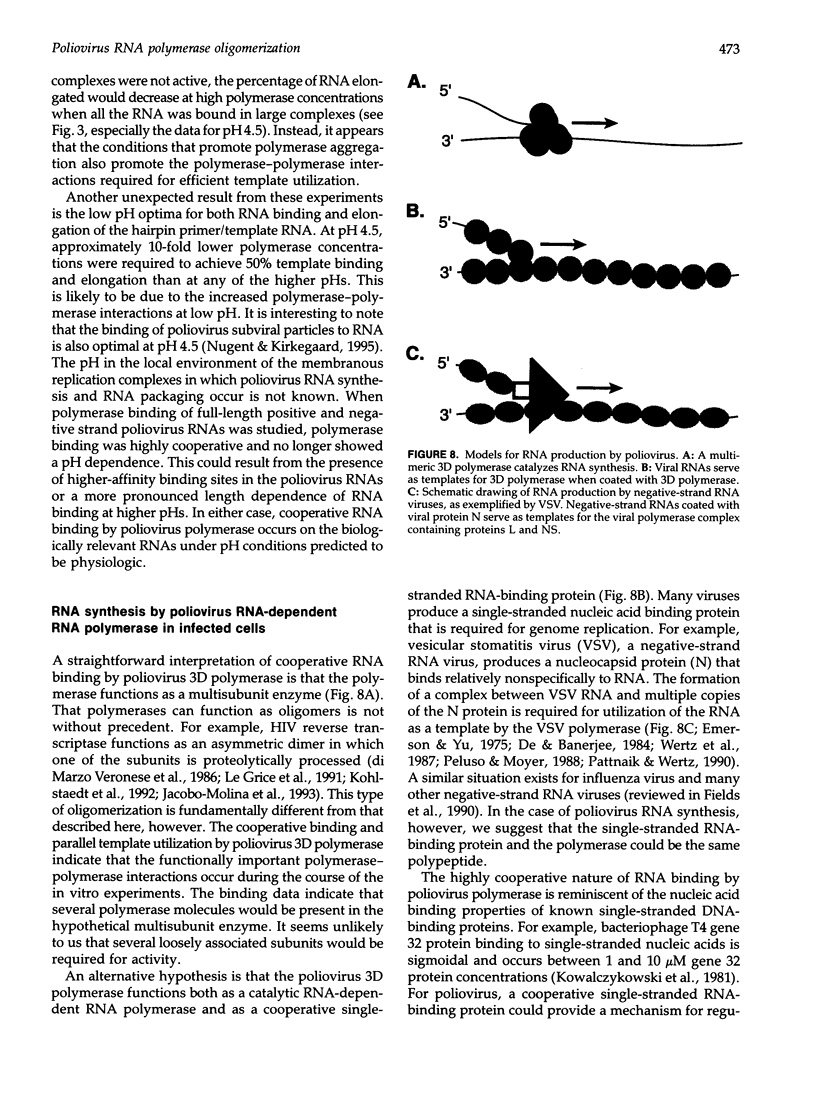
- Andino R., Rieckhof G. E., Achacoso P. L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5'-end of viral RNA. EMBO J. 1993 Sep;12(9):3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andrews N. C., Baltimore D. Purification of a terminal uridylyltransferase that acts as host factor in the in vitro poliovirus replicase reaction. Proc Natl Acad Sci U S A. 1986 Jan;83(2):221–225. doi: 10.1073/pnas.83.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N. C., Levin D., Baltimore D. Poliovirus replicase stimulation by terminal uridylyl transferase. J Biol Chem. 1985 Jun 25;260(12):7628–7635. [PubMed] [Google Scholar]
- Baron M. H., Baltimore D. In vitro copying of viral positive strand RNA by poliovirus replicase. Characterization of the reaction and its products. J Biol Chem. 1982 Oct 25;257(20):12359–12366. [PubMed] [Google Scholar]
- Baron M. H., Baltimore D. Purification and properties of a host cell protein required for poliovirus replication in vitro. J Biol Chem. 1982 Oct 25;257(20):12351–12358. [PubMed] [Google Scholar]
- Blumenthal T., Carmichael G. G. RNA replication: function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bruenn J. A. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991 Jan 25;19(2):217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. W., Richards O. C., Dmitrieva T. M., Agol V., Ehrenfeld E. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J Virol. 1993 Jun;67(6):3010–3018. doi: 10.1128/jvi.67.6.3010-3018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Zabel P., Baltimore D. Dependence of the activity of the poliovirus replicase on the host cell protein. Cell. 1980 Feb;19(2):423–429. doi: 10.1016/0092-8674(80)90516-4. [DOI] [PubMed] [Google Scholar]
- De B. P., Banerjee A. K. Specific interactions of vesicular stomatitis virus L and NS proteins with heterologous genome ribonucleoprotein template lead to mRNA synthesis in vitro. J Virol. 1984 Sep;51(3):628–634. doi: 10.1128/jvi.51.3.628-634.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond S. E., Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994 Feb;68(2):863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Yu Y. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975 Jun;15(6):1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc Natl Acad Sci U S A. 1977 Sep;74(9):3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez M. T., Eoyang L., August J. T. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968 Aug 10;219(5154):588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Harris K. S., Xiang W., Alexander L., Lane W. S., Paul A. V., Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994 Oct 28;269(43):27004–27014. [PubMed] [Google Scholar]
- Hayes R. J., Buck K. W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990 Oct 19;63(2):363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- Hey T. D., Richards O. C., Ehrenfeld E. Host factor-induced template modification during synthesis of poliovirus RNA in vitro. J Virol. 1987 Mar;61(3):802–811. doi: 10.1128/jvi.61.3.802-811.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson S. J., Konings D. A., Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3' terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993 Jun;67(6):2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis T. C., Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 1992 Aug;11(8):3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R. Characterization of the subunits of Q-beta replicase. Nature. 1970 Nov 7;228(5271):527–533. doi: 10.1038/228527a0. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kondo M., Gallerani R., Weissmann C. Subunit structure of Q-beta replicase. Nature. 1970 Nov 7;228(5271):525–527. doi: 10.1038/228525a0. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Lonberg N., Newport J. W., von Hippel P. H. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981 Jan 5;145(1):75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- Lama J., Paul A. V., Harris K. S., Wimmer E. Properties of purified recombinant poliovirus protein 3aB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J Biol Chem. 1994 Jan 7;269(1):66–70. [PubMed] [Google Scholar]
- Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991 Dec;10(12):3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A., Harris K. S., Paul A. V., Shin S. H., Mugavero J., Wimmer E. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J Biol Chem. 1994 Oct 28;269(43):27015–27020. [PubMed] [Google Scholar]
- Neufeld K. L., Richards O. C., Ehrenfeld E. Purification, characterization, and comparison of poliovirus RNA polymerase from native and recombinant sources. J Biol Chem. 1991 Dec 15;266(35):24212–24219. [PubMed] [Google Scholar]
- Nugent C. I., Kirkegaard K. RNA binding properties of poliovirus subviral particles. J Virol. 1995 Jan;69(1):13–22. doi: 10.1128/jvi.69.1.13-22.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Flanegan J. B. Measurement of poliovirus RNA polymerase binding to poliovirion and nonviral RNAs using a filter-binding assay. Nucleic Acids Res. 1988 Nov 11;16(21):10339–10352. doi: 10.1093/nar/16.21.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik A. K., Wertz G. W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990 Jun;64(6):2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. V., Cao X., Harris K. S., Lama J., Wimmer E. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J Biol Chem. 1994 Nov 18;269(46):29173–29181. [PubMed] [Google Scholar]
- Peluso R. W., Moyer S. A. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology. 1988 Feb;162(2):369–376. doi: 10.1016/0042-6822(88)90477-1. [DOI] [PubMed] [Google Scholar]
- Peters K., Richards F. M. Chemical cross-linking: reagents and problems in studies of membrane structure. Annu Rev Biochem. 1977;46:523–551. doi: 10.1146/annurev.bi.46.070177.002515. [DOI] [PubMed] [Google Scholar]
- Quadt R., Kao C. C., Browning K. S., Hershberger R. P., Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O. C., Martin S. C., Jense H. G., Ehrenfeld E. Structure of poliovirus replicative intermediate RNA. Electron microscope analysis of RNA cross-linked in vivo with psoralen derivative. J Mol Biol. 1984 Mar 5;173(3):325–340. doi: 10.1016/0022-2836(84)90124-4. [DOI] [PubMed] [Google Scholar]
- Roehl H. H., Semler B. L. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3' end of viral negative-strand RNA. J Virol. 1995 May;69(5):2954–2961. doi: 10.1128/jvi.69.5.2954-2961.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P. Role of 3'-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989 Jan;63(1):467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. H., Baltimore D. Polyadenylic acid on poliovirus RNA. II. poly(A) on intracellular RNAs. J Virol. 1975 Jun;15(6):1418–1431. doi: 10.1128/jvi.15.6.1418-1431.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T. A., Flanegan J. B. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J Virol. 1980 Sep;35(3):732–740. doi: 10.1128/jvi.35.3.732-740.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T. A., Rickles R. J., Flanegan J. B. Genome-length copies of poliovirion RNA are synthesized in vitro by the poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1982 Apr 25;257(8):4610–4617. [PubMed] [Google Scholar]
- Wertz G. M., Howard M. B., Davis N., Patton J. The switch from transcription to replication of a negative-strand RNA virus. Cold Spring Harb Symp Quant Biol. 1987;52:367–371. doi: 10.1101/sqb.1987.052.01.042. [DOI] [PubMed] [Google Scholar]
- Wimmer E., Hellen C. U., Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- Wong I., Lohman T. M. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. C., Tuschall D. M., Flanegan J. B. Poliovirus RNA-dependent RNA polymerase and host cell protein synthesize product RNA twice the size of poliovirion RNA in vitro. J Virol. 1985 May;54(2):256–264. doi: 10.1128/jvi.54.2.256-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haseth P. L., Uhlenbeck O. C. Interaction of Escherichia coli host factor protein with Q beta ribonucleic acid. Biochemistry. 1980 Dec 23;19(26):6146–6151. doi: 10.1021/bi00567a030. [DOI] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]