Abstract
FtsH, a AAA protease, associates with HflK/C subunits to form a megadalton complex that spans the inner membrane and extends into the periplasm of E. coli. How this complex and homologous assemblies in eukaryotic organelles recruit, extract, and degrade membrane-embedded substrates is unclear. Following overproduction of protein components, recent cryo-EM structures reveal symmetric HflK/C cages surrounding FtsH in a manner proposed to inhibit degradation of membrane-embedded substrates. Here, we present structures of native complexes in which HflK/C instead forms an asymmetric nautilus-like assembly with an entryway for membrane-embedded substrates to reach and be engaged by FtsH. Consistent with this nautilus-like structure, proteomic assays suggest that HflK/C enhances FtsH degradation of certain membrane-embedded substrates. The membrane curvature in our FtsH•HflK/C complexes is opposite that of surrounding membrane regions, a property that correlates with lipid-scramblase activity and possibly with FtsH’s function in the degradation of membrane-embedded proteins.
INTRODUCTION
AAA proteases sculpt the proteome to specific biological needs by using energy derived from ATP hydrolysis to unfold and degrade damaged or unneeded proteins (Baker and Sauer, 2012). Among AAA proteases, FtsH is unusual in being membrane anchored and thus positioned to degrade membrane proteins (Ito and Akiyama, 2005). FtsH homologs are widespread in eubacteria (Langklotz et al., 2012) and eukaryotic organelles of bacterial origin (Yi et al., 2022), and have been suggested as targets for anti-microbial (Hinz et al., 2011) and anti-malarial (Amberg-Johnson et al., 2017) therapeutics.
FtsH assembles into a homohexamer that is active in protein unfolding and degradation (Kihara et al., 1998; Tomoyasu et al., 1995). In Escherichia coli, each FtsH subunit consists of a transmembrane helix, a periplasmic domain, a second transmembrane helix, and a cytoplasmic AAA module and zinc-peptidase domain (Bittner et al., 2017; Ito and Akiyama, 2005) (Supplementary Figure 1). ATP hydrolysis within the AAA modules of FtsH powers substrate unfolding, extraction of membrane-embedded substrates from the lipid bilayer, and translocation of denatured polypeptides through an axial channel and into the peptidase chamber. FtsH degrades both membrane-bound and cytoplasmic proteins (Bittner et al., 2017), and is one of several AAA proteases in E. coli that degrade ssrA-tagged proteins resulting from tmRNA-mediated ribosome rescue (Herman et al., 2003). Disordered peptide segments in these substrates are engaged by the axial channel of the FtsH hexamer, allowing for specific recognition and subsequent mechanical unfolding and degradation (Bittner et al., 2017; Ito and Akiyama, 2005).
In E. coli, FtsH partners with two additional proteins – HflK and HflC, which contain SPFH domains, forming a large holoenzyme that extends across the inner membrane and into the periplasm (Saikawa et al., 2004) (Supplementary Figure 1). Similar super-complexes bearing SPFH domains also exist in eukaryotic organelles (Fu and MacKinnon, 2024). A recent cryo-EM structure of an FtsH super-complex from E. coli determined following overproduction of the protein components and the application of 4-fold symmetry (C4) during 3D reconstruction, revealed a cage consisting of 24 alternating subunits of HflK and HflC tightly enclosing four FtsH hexamers (Ma et al., 2022; Qiao et al., 2022). Paradoxically, however, this structure suggests that HflK/C prevents degradation of membrane-embedded substrates by insulating them from the FtsH proteolytic machinery.
Here, we present markedly different FtsH•HflK/C super-complex structures, which also contain 24 HflK/C subunits but only one or two FtsH hexamers. Notably, the HflK/C subunits in these structures form an asymmetric assembly, similar to a nautilus shell, with a passageway to the interior of the shell. These nautilus-like complexes were purified without protein overproduction using an affinity tag added to chromosomally encoded FtsH. Structures with similar topology were obtained after detergent solubilization or after detergent-free extraction using a nanodisc-forming polymer. The lipid domains in these structures display unexpected curvature, which correlates with enhanced rates of lipid scrambling. As such scrambling has been linked to a thinned membrane, this activity could aid FtsH in extracting and degrading membrane-embedded substrates. Additionally, we performed cell-wide proteomic experiments that revealed that several previously reported membrane-embedded protein substrates of FtsH are present at higher levels in the absence of HflK/C, supporting a model in which a nautilus-like FtsH•HflK/C holoenzyme degrades these substrates.
RESULTS
FtsH•HflK/C purification without protein overexpression.
We engineered an E. coli strain in which the chromosomal ftsH gene was modified to encode a C-terminal FLAG epitope and then used anti-FLAG antibody resin to purify tagged FtsH and associated HflK and HflC (Supplementary Figure 2A; see Methods). Addition of the FLAG tag did not slow E. coli growth at 37°C (Supplementary Figure 2B), despite FtsH protease activity being essential for cell viability (Herman et al., 1993). Thus, we inferred that the tag did not markedly affect activity in vivo. In preparation for single-particle cryo-EM, we used n-dodecyl-β-D-maltoside (DDM) or glyco-diosgenin (GDN) detergents to solubilize the FtsH•HflK/C complex prior to purification. Following DDM solubilization, the purified FtsH•HflK/C complex was active in ATP hydrolysis and ATP-dependent protein degradation (Supplementary Figure 2C-D).
Structural features of an asymmetric FtsH•HflK/C complex.
We refined a ~4.4 Å resolution cryo-EM structure of the DDM-solubilized complex without imposed symmetry, revealing an asymmetric HflK/C complex resembling a nautilus shell with two internal FtsH hexamers (Figure 1A-C; Supplementary Figures 3-4). A similar cryo-EM structure, albeit at lower resolution, was obtained when the complex was solubilized with the detergent GDN (Supplementary Figures 5), indicating that the nautilus-like structure was not unique to a specific detergent. Moreover, this nautilus-like structure was not the result of preferential orientations or preferential positioning at the air-water interface, as the 3D-FSC (Tan et al., 2017) measure of sphericity was 0.89 with diverse projection angles observed (Supplementary Figure 4C-D). Furthermore, tomographic reconstructions revealed random particle orientations on the EM grid (Supplementary Figure 6).
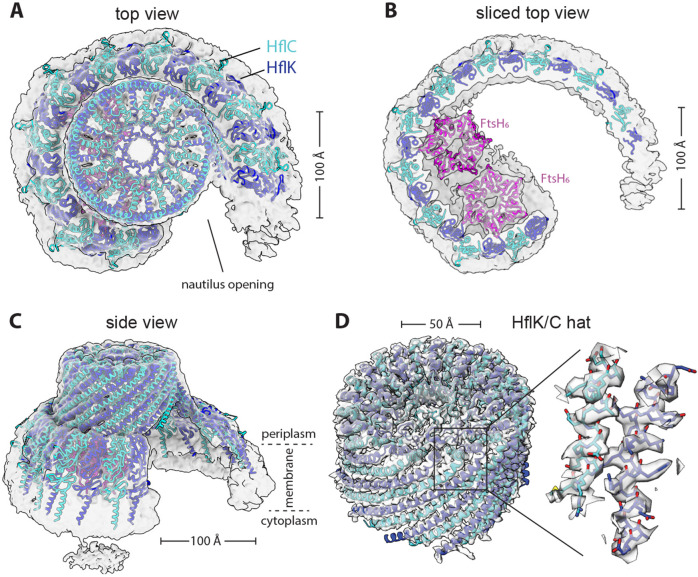
Figure 1. Nautilus-like structure of a FtsH•HflK/C complex.
(A) Density map (unsharpened) and cartoon models of HflK (blue) and HflC (cyan) viewed from top. Nautilus opening into the HflK/C chamber noted. Map was resolved to a GS-FSC resolution of ~4.5Å (B) A sliced top view of the complex highlighting two FtsH hexamers (pink) interacting with SPFH domains of HflK within the nautilus chamber. (C) A side view of the nautilus super-complex, as shown in (A). Approximate location of the membrane depicted with periplasmic and cytoplasmic faces of the inner membrane noted. (D) A hat-like portion of the structure locally refined to ~3.5Å GS-FSC resolution, depicted as in (A). This map allowed for clear identification of HflK and HflC. The inset displays is a sharpened map and the model built using this map, highlighting bulky residues in helical regions of HflK and HflC.
HflK and HflC share sequence and structural homology and were difficult to distinguish in the 4.4-Å density map of the DDM-solubilized complex. However, after signal subtraction and local refinement in C1 (see Methods), we refined the periplasmic portion of the nautilus structure farthest from the membrane, which resembles a hat, to 3.5-Å resolution (Supplementary Figure 7-8). This structure allowed visualization of side chains and unambiguous assignment of ~110 C-terminal residues of 24 alternating HflK and HflC subunits (Figure 1D). The corresponding hat region of the C4-symmetric HflK/C structure (pdb 7WI3) (Qiao et al., 2022) aligned to the asymmetric nautilus hat with an RMSD of 2.3 Å for the 2,545 common Cα positions. SPFH domains, present in each HflK and HflC subunit, formed the membrane-proximal sides of the nautilus chamber, but only 21 of 24 these domains could be modeled (Figures 1-2). The entryway into the nautilus chamber was located near the ‘missing’ SPFH domains, which presumably adopted multiple conformations or were proteolytically truncated. The final model, which had good geometry (Table 1), included residues 79-355 for most HflK subunits, residues 1-160 and 191-329 for most HflC subunits, and residues 31-97 of the periplasmic domains of two FtsH hexamers.
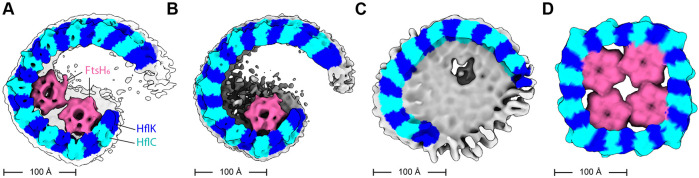
Figure 2. HflK/C structures with different numbers of FtsH hexamers.
Density maps were low-pass filtered to 10 Å and colored using the underlying atomic model (HflK – blue; HflC – cyan, FtsH – pink; unassigned membrane or detergent – grey). (A) Two FtsH hexamers per super-complex as refined in Supplementary Figure 10. (B) One FtsH hexamer per super-complex as refined in Supplementary Figure 9. (C) FtsH-free super-complex affinity-purified from cells overexpressing HflC-FLAG and HflK, and refined as depicted in Supplementary Figures 11-12. (D) C4-symmetric structure with four FtsH hexamers (Qiao et al., 2022).
Table 1.
Cryo-EM data collection, processing, model building, and validation statistics.
| Sample and data deposition information | ||||||
|---|---|---|---|---|---|---|
| Sample name | FtsH•HflK/C map a (DDM extracted) | FtsH•HflK/C hat-like map (locally refined) | FtsH•HflK/C map I | FtsH•HflK/C map II | HflK/C | FtsH•HflK/C (carboxy-DIBMA) |
| Description | Primary map analyzed | Locally refined map a focused on HflK/C ‘hat’ | Classified map a with single FtsH6 | Classified map a with two FtsH6 | HflK/C overexpressed | Detergent-free solubilized sample |
| PDB code | 9CZ2 | 9CZ1 | n/a | n/a | n/a | n/a |
| EMDB ID | 46057 | 46056 | 46061 | 46059 | 46062 | 46058 |
| EMPIAR code | to be deposited | |||||
| Defocus range (μm) | −0.05 to −1.75 | −0.5 to −2 | −0.30 to 1.75 (1); −0.75 to −2.0 (2) | |||
| Micrographs collected (#) | 25,179 | 16,055 | 11,899 (1); 32,907 (2) | |||
| Pixel size (Å) | 0.654 | 0.654 | 0.654 | |||
| Map reconstruction | ||||||
| Image processing package | cryoSPARC | |||||
| Total extracted particles | 334,256 | 334,256 | 295,185 | 47,449 | ||
| Final particle count | 184,019 ∣ 184,019 | 17,473 | 76,491 | 230,567 | 24,011 | |
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 | |
| Resolution (Å) | ||||||
| GS-FSC (0.143) | ||||||
| unmasked | 7.0 | 7.0 | 12 | 8.0 | 9.7 | 15 |
| loose | 5.6 | 3.7 | 7.1 | 6.0 | 9.1 | 14 |
| tight | 4.4 | 3.5 | 6.6 | 4.5 | 7.7 | 12 |
| 3D-FSC sphericity (out of 1.0) | 0.89 | 0.82 | 0.712 | 0.906 | 0.926 | 0.870 |
| Model composition and refinement | ||||||
| Number of atoms | 114,335 | 40,604 | no models built | |||
| Protein residues | 7,130 | 2,556 | ||||
| Refinement package | Phenix and Coot | |||||
| Map-to-model cross correlation | ||||||
| masked | 0.69 | 0.77 | ||||
| unmasked | 0.72 | 0.78 | ||||
| RMSD bond length (Å) [#>4σ] | 0.003 [0] | 0.003 [0] | ||||
| RMSD bond angles (°) [#>4σ] | 0.702 [0] | 0.664 [0] | ||||
| MolProbity score | 0.88 | 0.91 | ||||
| Clash score | 1.43 | 2.48 | ||||
| C-beta outliers (%) | 0.0 | 0.0 | ||||
| Rotamer outliers (%) | 0.03 | 0.0 | ||||
| Ramachandran favored (%) | 99.9 | 100 | ||||
The nautilus chamber in our structure contained two hexamers composed of FtsH periplasmic domains, with one hexamer better resolved than the other. In their closed C4-symmetric structure, Qiao et al. identified contacts between Arg191 of HflK and residues 62-64 of the periplasmic domain of FtsH. Although the resolution of our asymmetric nautilus structure was not sufficient to confidently assign these exact contacts, the backbones in both structures were positioned similarly (Figures 1B, 2). Because the tightly closed HflK/C cage in the C4-symmetric structure (Qiao et al., 2022) contained four FtsH hexamers, whereas our open nautilus chamber contained fewer FtsH hexamers, we hypothesized that chamber curvature might be proportional to the number of internal FtsH hexamers. To test this hypothesis, we performed focused classification, using a mask encompassing the periplasmic hexamer of FtsH with lower occupancy (Supplementary Figure 3). After 3D-classification and refinement of the classified particles, assemblies with either one or two FtsH hexamers were evident (Supplementary Figure 9-10). Comparison of these maps (Figure 2A-B) revealed no significant structural rearrangements in the nautilus shell, indicating that chamber curvature does not simply increase with the number of enclosed FtsH hexamers. Moreover, when we overproduced HflK and HflC prior to affinity purification via a FLAG-tag on HflC, the resulting cryo-EM structure (Figure 2C, Supplementary Figures 11-12) had a nautilus-like HflK/C chamber lacking FtsH density, but with curvature similar to our structures with one or two FtsH hexamers. In each instance, our maps were clearly distinct from the C4-symmertrized structure from Qiao et al. (Figure 2D).
We next searched for structural heterogeneity in our DDM-solubilized FtsH•HflK/C dataset using cryoDRGN (Kinman et al., 2023; Sun et al., 2023; Zhong et al., 2021). This analysis revealed the expected nautilus-like structures with either one or two FtsH hexamers and additional differences in the width of the entryway into the nautilus chamber (Supplementary Movie 1), consistent with flexibility in the SPFH domains of the terminal HflK/C subunits. Notably, however, closed HflK/C structures or complexes with four FtsH hexamers were not observed.
Unusual curvature of the membrane domain of FtsH•HflK/C.
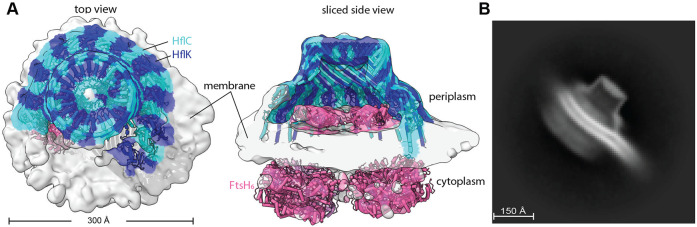
The density map of DDM-solubilized FtsH•HflK/C showed the approximate positions of lipid head groups in the bilayer (Supplementary Figure 4), but individual lipid molecules could not be resolved or modeled. Notably, however, the bilayer curved inward within the complex, whereas the bacterial inner membrane as a whole curves outward. This unusual membrane geometry was not a function of detergent solubilization of the complex, as similar curvature was observed in a lower-resolution (~12Å) 3D reconstruction of the affinity purified FtsH•HflK/C complex following extraction with the nanodisc-forming polymer Carboxy-DIBMA, and native lipids (Figure 3A, Supplementary Figures 13-14). Notably, a 2D class average side view of these particles showed pronounced inward membrane curvature within the complex and reversal of this curvature as the membrane extended beyond the embedded FtsH•HflK/C complex (Figure 3B). Density for the cytoplasmic portions of both FtsH hexamers in the Carboxy-DIBMA map was far stronger than in the higher-resolution map of the DDM-solubilized complex, which may reflect the presence of native lipids or other differences in the protocols used to purify the complexes.
Figure 3. Nautilus-like FtsH•HflK/C structure isolated with native lipids via detergent-free extraction.
(A Density map highlighting the nautilus-like FtsH•HflK/C structure determined after affnity purification following detergent-free extractoin using Carboxy-DIBMA (see Methods). Map is color-coded (HflK, blue; HflC cyan; FtsH, pink) using a docked atomic model and is displayed from the top (left) and as a sliced side view (right). Key structural elements and scale bar noted. (B) 2D-class average of the Carboxy-DIBMA-extracted sample, highlighting membrane reshaping, as evidenced by the belt-like membrane structure that exhibits dual curvature as it passes through and extends beyond the particle.
FtsH•HflK/C enhances lipid scrambling.
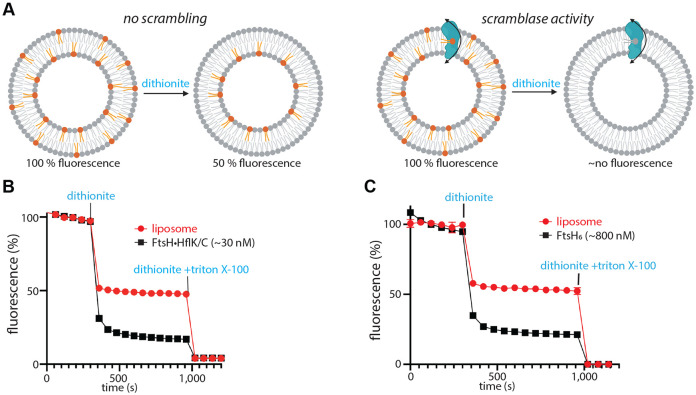
Unusual membrane curvature often correlates with enhanced rates of polar lipid head group flipping from one side of a bilayer to the other (Devaux et al., 2008; Jarsch et al., 2016). This activity can be assayed by reconstituting proteoliposomes with fluorescent NBD-labeled lipids (Ghanbarpour et al, 2021; Ploier & Menon, 2016), which orient randomly in proteoliposomes, with approximately half of the fluorescent lipids initially in the inner leaflet and half in the outer leaflet. Treatment with dithionite, which does not penetrate liposomes, quenches the fluorescence of outward-facing NBD groups, thereby reducing the fluorescence to ~50% of the initial level (Figure 4A, left). If proteins that catalyze lipid scrambling are present in the membrane, dithionite treatment can further reduce fluorescence, as NBD-labeled lipids initially facing the lumen can be translocated to the outside, where they are exposed to dithionite quenching (Figure 4A, right). Using this assay, we found that FtsH•HflK/C and FtsH catalyze lipid scrambling (Figure 4B-C). Although this activity did not require ATP hydrolysis by FtsH, we postulate that it may help this ATP-fueled protease to extract membrane proteins from the lipid bilayer during degradation (see Discussion).
Figure 4. FtsH-catalyzed lipid scramblase activity.
(A) Schematic representation of the scramblase activity assay, with expected fluorescence level in the absence (left) or presence (right) of enzymatic lipid scrambling activity. Scramblase activity assay of the FtsH•HflK/C complex (B) or isolated FtsH6 (C) using NBD-PE as a substrate. Note that the reported protein concentrations do not consider potential differences in proteoliposome reconstitution efficiency between samples, and that not all liposomes necessarily contain protein. Control reactions to asssess the FtsH-dependence of the measured scramblase activity include: 1) absense of scramblase activity using detergent-solubilized GlpG membrane protein (Ghanbarpour et al., 2021); and 2) NBD-glucose and BSA-back-extraction assays demonstrating that the measured scramblase activity was not a result of partially disrupted, leaky liposomes (Supplementary Figure 15).
HflK/C impacts intracellular protein levels.
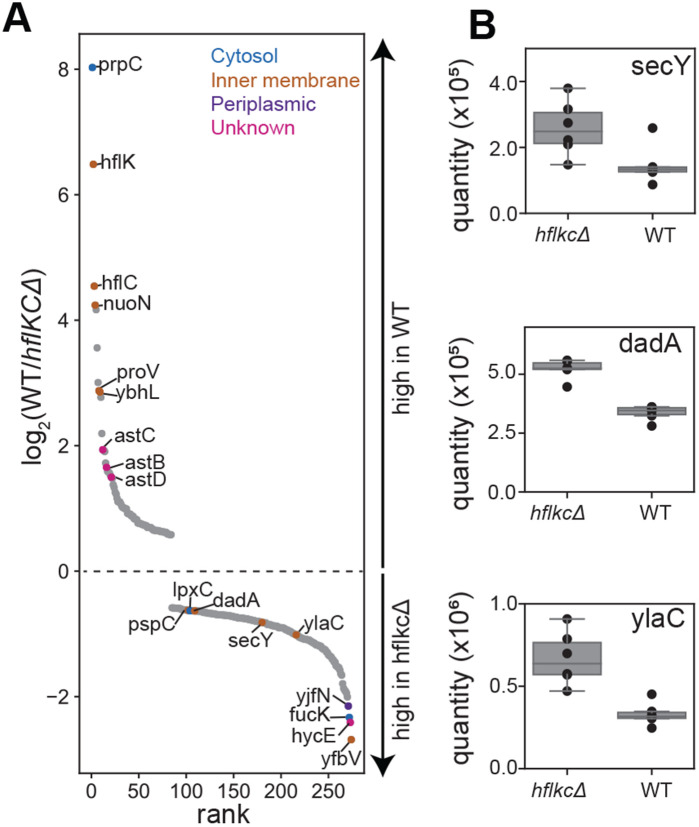
To investigate how HflK/C affects E. coli protein levels, we used mass spectrometry to quantify the steady-state abundance of ~2,250 proteins in wild-type cells or mutant cells lacking HflK/C (ΔhflK/C) (Figure 5). Of these proteins, ~88% showed no significant differences, ~8% were more abundant in ΔhflK/C cells, and ~4% were less abundant in these mutant cells. We considered that FtsH•HflK/C could degrade proteins in the ‘more abundant’ category faster than FtsH alone, and that HflK/C could restrict degradation of proteins in the ‘less abundant’ category. However, other possibilities include indirect effects of hflK/C genetic deletion, such as an impact on FtsH-driven degradation of proteins that regulate the transcription, translation, or turnover of these differentially abundant proteins. Notably, proteins in the ‘more abundant’ category included the membrane proteins DadA, PspC, SecY, and YlaC, which are known to be degraded by FtsH (Arends et al., 2016; Singh and Darwin, 2011; Westphal et al., 2012). To identify proteins that directly interact with FtsH, we affinity-purified FtsH from wild-type or ΔhflK/C cells, and analyzed the eluate by mass spectrometry. Remarkably, some of the differentially abundant proteins identified above, including DadA and AstC, also exhibited HflK/C-dependent association with FtsH (Extended Data Table 1).
Figure 5. Steady-state protein abundance measurements in wild-type and ΔhflK/C cells.
(A) Semi-quantitative proteomic measurements of protein abundance in wild-type or ΔhflK/C strains of E. coli. Proteins (n=274) exhibiting statistically significant changes (p<0.01) across 6 replicates are plotted following the measured rank order abundance change between cell types. A subset of proteins are labeled by gene name and colored according to their annotated localization. Among the highlighted ΔhflK/C-enriched proteins are the putative FtsH substrates lpxC, dadA, secY, and ylaC. (B) Signal intensity (quantified using precursor ion intensities with Spectronaut®) compared between wild-type and ΔhflK/C cells. Inner-quartile ranges are depicted as box plots, with dots indicating the abundance measured in each of 6 replicates. Changes in protein levels were statistically significant, with multiple hypothesis testing corrected q-values of 7.9*10−5 (secY), 3.6*10−11 (dadA), and 8.2*10−07 (ylaC).
DISCUSSION
In E. coli, the SPFH-family HflK and HflC proteins interact with the AAA FtsH protease to form a large holoenzyme (Kihara et al., 1996; Saikawa et al., 2004). The first cryo-EM structures, which were solved after protein overproduction using C4-symmetry in the reconstruction process, showed HflK/C subunits forming a tight symmetric cage around four FtsH hexamers (Ma et al., 2022; Qiao et al., 2022). In these structures, the transmembrane helices of HflK/C completely surround the internal FtsH hexamers and would appear to insulate FtsH from interactions with potential membrane-embedded substrates. By contrast, following purification of FtsH•HflK/C without overexpression, the nautilus-like cryo-EM structures presented here, which were determined without imposed symmetry, revealed an HflK/C chamber containing one or two FtsH hexamers and an opening that could allow membrane proteins to enter and be engaged by FtsH. Although it is possible that a population of cellular FtsH•HflK/C corresponds to the symmetric caged structure, measured cellular protein stoichiometries in E. coli (Li et al., 2014) preclude such structures being uniformly present. Specifically, the abundances of FtsH, HflK, and HflC in serveral growth conditions are only sufficient for an average of ~2.3 FtsH hexamers for every 24 subunits of HflK/C (Li et al., 2014). Moreover, a model in which cells contain approximately equal amounts of free FtsH hexamers and sets of four FtsH hexamers tightly caged by HflK/C is inconsistent with experiments showing that the vast majority of FtsH co-sediments in a large complex with HflK/C, with very few free FtsH hexamers observed (Saikawa et al., 2004). Our affinity isolation experiments were also consistent with this observation, as the stoichiometry of the components in our purified FtsH•HflK/C sample (Supplementary Figure 2) was similar to that in cells. Thus, we posit that the nautilus-like FtsH•HflK/C structures described here likely account for a major fraction of holoenzyme complexes in the cell. Notably, most bacteria that encode FtsH also encode HflK and HflC homologs, and mitochondrial relatives of FtsH associate with SPFH family proteins (Fu and MacKinnon, 2024; Wessel et al., 2023), raising the possibility that such super-complexes are present in a variety of organisms.
Structurally, one would expect the previously reported symmetric HflK/C enclosure to prevent FtsH degradation of membrane-embedded substrates, whereas the nautilus-like structure, with its open portal, could enhance degradation of some membrane proteins and slow degradation of others. For example, interactions between membrane-embeddd proteins and structural elements of HflK/C within the nautilus chamber could increase substrate residence times, thereby making the engagement of degron sequences by the cytoplasmic proteolytic machinery of FtsH more probable. Supporting this idea, our proteomic experiments showed that genetic deletion of hflK/C increased the abundance of several membrane proteins that were previously shown to be substrates of FtsH (Arends et al., 2016; Rei Liao and van Wijk, 2019; Westphal et al., 2012) (Figure 5). Additional membrane and soluble proteins also had different steady-state levels in wild-type and ΔhflK/C cells, but whether these proteins are direct substrates or their levels are affected by another FtsH•HflK/C substrate remains to be determined. Conversely, the entryway into the HflK/C chamber, which spans ~70 Å, could prevent the entry and degradation of sufficiently large membrane-protein complexes, with the HflK/C nautilus negatively regulating FtsH activity. Finally, HflK/C appears to influence the associated lipid domain and membrane localization of FtsH (Wessel et al., 2023), which could be important in determining the local pool of membrane-embedded substrates subject to FtsH-mediated degradation.
We observed reverse-membrane curvature in our FtsH•HflK/C complexes, which was most evident in the detergent-free nanodisc samples (Figure 3). Reverse membrane curvature, coupled with a potentially different lipid composition within the FtsH•HflK/C domain (Browman et al., 2007; Wessel et al., 2023), might facilitate the recruitment of certain membrane-embedded substrates or adaptors into the FtsH•HflK/C complex. Indeed, in eukaryotic cells, altered membrane curvature is often seen within lipid rafts, which can act as cellular reaction centers within the plasma membrane (Sadeghi et al., 2014). FtsH and FtsH•HflK/C each catalyze the rate at which lipids change orientation within membrane bilayers. A simple way in which this could occur is if FtsH and the complex were to thin parts of the associated membrane, as this would reduce the energetic penalty for passage of the head group through the hydrophobic interior of the membrane. In mitochondria, FtsH homologs regulate lipid metabolism (Chiba et al., 2006; Löser et al., 2021), which may depend on enhanced lipid flipping and downstream processes. Membrane thinning could also assist in the degradation of membrane-embedded substrates by FtsH. Here, we envision that the energetic cost of extracting a membrane protein from the bilayer during degradation would be reduced if the membrane were thinner, an idea that has been advanced to explain the CDC48-dependent degradation of ERAD-L substrates in eukaryotes (Wu et al., 2020). Taken together, the unique structural features of this super-complex, including the asymmetric nautilus-like assembly and its impact on the local membrane curvature may support FtsH’s critical role in maintaining proteostasis at the membrane.
MATERIALS AND METHODS
FtsH•HflK/C purification.
A C-terminal FLAG-tagged variant of FtsH was introduced into E. coli BL21(DE3) at the endogenous FtsH locus using the no-SCAR (Scarless Cas9 Assisted Recombineering) system (Reisch & Prather, 2017). Briefly, the spacer sequence GTCGCCTAACTGCTCTGACA targeting the C-terminus of FtsH was closed into the temperature sensitive pKDsg-XXX plasmid, and introduced into BL21(DE3) cells containing the pCas9cr4 plasmid. λ-Red recombineering (Datsenko & Wanner, 2000) was induced, followed by transformation with a 500 bp double-stranded gene block (IDT) containing a mutation in the PAM sequence that appended the sequence coding for “SGSGDYKDDDK” to the C-terminus of FtsH. The chromosomal mutation was selected for by inducing Cas9 and subsequently verified by amplifying the region using PCR and DNA sequencing. BL21(DE3) cells expressing this C-terminal FLAG-tagged FtsH were grown overnight in 50 mL of Luria broth (LB), transferred to a 1:1 LB:2xYT medium mixture, and grown for an additional 20 h to an A600 of ~5.5. Cells were harvested by centrifugation at 4,500 rpm for 30 min and resuspended in buffer A [20mM HEPES (pH 7.5), 400mM NaCl, 100mM KCl, 10% glycerol, 1mM DTT] before flash freezing in liquid N2 for storage at −80°C.
For cell lysis, resuspended cells were sonicated (QSonica) on ice using 30% power with cycles of 10 s on and 30 s off for a total duration of 4 min. Following sonication, cells were centrifuged at 12,100 x g for 20 min in a JL25.50 rotor. The supernatant was isolated and then centrifuged at 105,000 x g for 1 h using a 45 Ti rotor. The pelleted membrane was resuspended in 5 mL of buffer A and disrupted using a Dounce homogenizer. Detergent n-dodecyl-β-D-maltoside (DDM) was added to a final concentration of 1%, followed by platform rotation at 4°C for 1 h. The mixture was centrifuged for 20 min at 35,000 rpm using a TLA 100.3 rotor, incubated with anti-Flag M2 resin overnight, washed with a buffer containing 0.03% DDM, and finally eluted with 0.25 mg/mL Flag peptide (APEXBio) according to the manufacturer’s directions. The same protocol was used for glyco-diosgenin (GDN) sample preparation, except 2% GDN was used for membrane solubilization and 0.03% GDN was employed for the wash and elution steps.
The overexpressed HflK/C sample was prepared using the petDUET plasmid as previously reported (Ma et al., 2022). Briefly, HflC followed by a C-terminal Flag-tag was cloned into the first multicloning site, and HflK with an N-terminal octa-histidine tag was cloned into the second site. BL21 (DE3) cells harboring the plasmid were grown at 37°C from an overnight cell culture to an OD of ~1. Expression was induced by the addition of 0.2mM IPTG and continued overnight at 18°C. Purification was performed similarly to the procedure described for the endogenously expressed FtsH-HflK/C solubilized in DDM.
Carboxy-DIBMA extraction.
Carboxy-DIBMA was synthesized analogously to Sulfo-DIBMA as described (Glueck et al., 2022). Membranes, isolated as described above, were resuspended in buffer B [50mM Tris-HCl (pH 8.0), 100mM NaCl, 5mM MgSO4, 100μM ZnCl2, and 3mM β-mercaptoethanol], to a final concentration of approximately 35 mg/mL. To this suspension, 22 mg/mL of Carboxy-DIBMA solution and 1 mM ATP were added, bringing the total volume to 5 mL. The mixture was gently rotated overnight at 4°C to facilitate solubilization. Subsequently, samples were centrifuged at 30,000 x g, yielding a clear supernatant. This supernatant was incubated with anti-FLAG M2 magnetic beads for 2 h at 4°C with gentle rotation. After incubation, the sample was centrifuged at 300 x g, the supernatant was carefully removed with a magnet, and beads were washed with 15 mL of buffer B. Finally, the sample was eluted using 0.5 mg/mL FLAG peptide. The eluted sample was concentrated, analyzed, and quantified after SDS-PAGE, using bovine serum albumin as a standard.
Cryo-EM sample preparation.
DDM- or GDN-solubilized samples were prepared by applying 2.5 μL of ~0.75 mg/mL of FtsH•HflK/C complex onto homemade graphene-supported 200-mesh Quantifoil 2/1 copper grids (Grassetti et al., 2023), pre-treated with UV/ozone for 7 min using a Bioforce UV/ozone cleaner. Sample-loaded grids were blotted for 4 s with a blot force of +4 at 6°C and 95% relative humidity using a FEI Vitrobot Mk IV instrument. The same concentration of the overexpressed HflK/C sample was used for grid preparation. However, in this case, 2-nm carbon-supported 200-mesh Quantifoil 2/1 copper grids, which had been glow-discharged for 20 s in an easiGlow glow discharger (Pelco) at 15 mA, were utilized. For the Carboxy-DIBMA-extracted sample, a concentration of ~0.25 mg/mL of the FtsH•HflK/C complex was applied to 2-nm carbon-supported 200-mesh Quantifoil 2/1 copper grids, also glow-discharged for 20 s in an easiGlow glow discharger (Pelco) at 15 mA, and the sample was vitrified as above.
Cryo-EM data collection.
For the DDM-solubilized FtsH•HflK/C complex, 25,179 movies were collected with EPU using aberration-free image shift (AFIS) and hole-clustering method on a Titan Krios G3i with an acceleration voltage of 300 kV and magnification of 130,000x, detected in super-resolution mode on a Gatan K3 detector for an effective pixel size of 0.654 Å (binned by 2). Movies were collected as 40 frames with a defocus range from −0.05 to −1.75 μm and a total exposure per specimen of 46.1 e−/Å2. For the DDM-solubilized and overexpressed HflK/C complex, movies were collected as 40 frames with a defocus range from −0.5 to −1.75 μm and a total exposure per specimen of 47.2 e−/Å2 using AFIS hole clustering. For the Carboxy-DIBMA extracted sample, movies (11,989) were collected as 40 frames with a defocus range from −0.30 to −1.75 μm and a total exposure per specimen of 44.6 e−/Å2 using AFIS hole clustering. An additional data collection session on this sample produced 32,907 movies, collected with a defocus range from −0.75 to −2.0 μm and a total exposure per specimen of 44.4 e−/Å2 using AFIS hole clustering.
Cryo-EM pre-processing and particle picking.
For the DDM-solubilized FtsH•HflK/C complex, data processing was performed using cryoSPARC (v4.0) and default parameters unless noted (see Supplementary Figure 3). Raw movies (25,179) were pre-processed using ‘Patch motion correction’, and ‘Patch CTF estimation’. Particles (73,171) were picked using the ‘Blob-picker’ tool applied to 1,000 micrographs selected at random. Particles were extracted (box size 640px, Fourier cropped to 360px) and classified using the ‘2D classification’ utility. The entire set of micrographs were then picked with the ‘Template picker’ tool, using a single class from 2D classification. Particles were extracted (box size 640px, Fourier-cropped to 360px) and subjected to three rounds of 2D classification, resulting in the selection of 334,256 particles as a preliminary stack.
For the DDM-solubilized overexpressed HflK/C complex, data processing was performed using cryoSPARC (v4.0) and default parameters unless noted (see Supplementary Figure 11). Raw movies (16,055) were pre-processed and particles picked as described above, except that the extraction-box-size was 900px (Fourier cropped to 300px). After four rounds of 2D classification, 295,185 particles were selected as a preliminary stack.
For the FtsH•HflK/C complex extracted with Carboxy-DIBMA, data processing was carried out using cryoSPARC (v4.0) with default settings unless noted (see Supplementary Figure 13). Raw movies were collected over two sessions. Data from these sessions were pre-processed individually using ‘Patch motion correction’ and ‘Patch CTF estimation’. Particles were separately extracted (box size of 1,000px, Fourier cropped to 360px). After several rounds of 2D classification of particles from the isolated datasets, the particles were pooled and subjected to two additional rounds of 2D classification, resulting in the final particle stack of 47,449 particles.
Ab initio reconstruction, global refinement, and model building.
For the FtsH•HflK/C complex solubilized in DDM, ‘Ab-initio’ reconstruction using two classes was performed, with a resulting single class model and particle stack (184,019 particles) selected for subsequent ‘Homogenous refinement’ followed by ‘Non-uniform refinement’. A mask was then created in ChimeraX (Pettersen et al., 2021) that only included the periplasmic portion of HflK/C and FtsH, excluding the cytoplasmic domain of FtsH, which exhibited substantial flexibility. Using this mask, ‘Local refinement’ was performed, leading to a ~4.4 Å GS-FSC resolution map we termed “map a”. To separate single vs. double FtsH hexamers within the HflK/C assembly, a mask was generated that encompassed the FtsH periplasmic domain with a lower occupancy. Using this mask, ‘3D classification’ (20 classes) was performed. Based on visual inspection, particles assigned to maps bearing one FtsH hexamer were pooled and subjected to ‘Local refinement’ using the original mask, producing “map I”. A parallel operation was performed on particles assigned to maps bearing two hexamers, resulting in “map II”.
To generate a higher-resolution map for the ‘hat’ portion of the complex a signal subtraction and local refinement strategy was employed (see Supplementary Figure 7). Briefly, two masks were generated using map a - one encompassing on the hat portion, and another containing the remainder of map a, which was used for signal subtraction after first locally aligning the particles using the non-hat mask using the ‘Local refinement’ utility. Next, the signal-subtracted particles and the mask around the hat portion were used to generate an initial volume using the ‘Homogeneous reconstruct only’ utility followed by a ‘Local refinement’ using the same mask. After application of the ‘Global CTF refinement’ followed by ‘Local CTF refinement’, the final map reached ~3.5 Å GS-FSC resolution.
For the overexpressed HflK/C complex solubilized in DDM, ‘Ab-initio reconstruction’ using three classes was performed, and one class was selected (106,858 particles). ‘Homogeneous refinement’ followed by ‘Non-uniform refinement’ were then employed, producing a map that was used as an initial model (after low-pass filtering) in a final round of ‘Non-uniform refinement’ followed by ‘Local refinement’ with the original particle stack (230,567 particles). After conducting both global and local CTF refinements, the final map achieved a GS-FSC resolution of ~9.8 Å.
For the FtsH•HflK/C complex extracted with Carboxy-DIBMA, ‘Ab-initio reconstruction’ was carried out on the 47,449 particles using two classes. Of these, 24,011 particles were selected for ‘Non-uniform refinement’, followed by global and local CTF refinement, and an additional round of ‘Local refinement.
Model building was performed using a combination of ChimeraX-1.3 (Pettersen et al., 2021), Coot (0.9.4) (Casanal et al., 2020), and Phenix (1.14) (Liebschner et al., 2019). Local resolution was estimated by cryoSPARC implementation of monoRes (Vilas et al., 2018).
Tilt-series acquisition.
The GDN-solubilized FtsH•HflK/C sample was used for tomographic data collection. A total of 18 tilt series were acquired on a Titan Krios G3i microscope operating at 300kV. Movies were collected at a nominal magnification of 64,000x on a BioQuantum K3 detector operating in super-resolution mode, and were pre-binned at collection time to a physical pixel size of 1.36 Å, with 6 frames per movie. Tilt series were collected following a dose-symmetric scheme from −58° to 58° in 2° increments, beginning from 0°, and each tilt series was collected at a nominal defocus stepped between −3 μm and −5 μm. The total electron dose per tilt series was 117.2 e−/Å2. The energy-filter slit width was 20 eV during acquisition, and the zero-loss peak was refined between each tilt movie acquisition.
Tomogram reconstruction and sub-tomogram averaging.
Tomographic data processing generally followed Powell et al., 2024, with the following modifications. Tilt movies were aligned and initial CTF parameters estimated in Warp (v1.0.9) (Tegunov and Cramer, 2019). Tilt-series stacks were exported and aligned in IMOD (v4.11.12) using patch tracking (Mastronarde and Held, 2017). Tilt series were re-imported into Warp with alignment parameters and CTF parameters were fit using each full-tilt series. Tomograms were reconstructed and deconvolved in Warp using a pixel size of 10 Å/px. All particles were manually picked using Cube (n=1,578 particles). Particles were exported from Warp with pixel size of 8 Å/px and box size of 64 px3. RELION (v3.1.3) (Zivanov et al., 2018) was used for initial model generation and subsequent particle refinement with C1 symmetry, reaching an unmasked resolution of 24.4 Å. Refined particles were imported to M (v1.0.9) (Tegunov et al., 2021) and subjected to iterative refinement of image warp, volume warp, particle pose, and stage angle parameters, ultimately reaching a masked resolution of 17.9 Å. Particles were re-extracted from M with pixel size of 8 Å/px and a box size of 64 px3, re-refined in RELION, re-imported to M, and re-refined in M, reaching a final resolution of 17.3 Å. Copies of this final reconstruction were mapped back in the source tomogram and visualized with ChimeraX using the subtomo2chimerax functionality implemented in tomoDRGN (Powell and Davis, 2024).
Mass spectrometry.
For steady-state protein comparisons between wild-type and ΔhflK/C E. coli strains, cells were grown in LB and harvested via centrifugation at 4°C at an A600 of ~1. Cell pellets were frozen at −80°C for future use. Thawed cell pellets were washed with buffer MS1 [20mM Tris-HCl (pH 7.5), 100mM NH4Cl, 10mM MgCl2, 0.5mM EDTA, and 6mM β-mercaptoethanol], followed by the addition of SDS (10% final concentration) and 0.25 U/μL benzonase (EMD Millipore). After incubation for 10 min at room temperature, lysates were cleared via centrifugation at 21,000 x g for 8 min. DTT (final concentration of 20 mM) was introduced to the lysate, followed by heating at 65°C to reduce disulfide bonds. After cooling to 30°C, iodoacetamide (final concentration of 40 mM) was added for 30 mins to alkylate free cysteines. Subsequent acidification, column preparation and sample binding to S-Trap™ micro (≤ 100 μg) spin columns and column washing followed the manufacturer’s protocol (ProtiFi). A trypsin solution (Promega Inc.) was combined with 200 μL of digestion buffer and applied to the spin columns containing bound proteins followed by a 1 h incubation at 42°C. Peptides were eluted using a sequential addition of 50 mM TAEB and 0.2% (v/v) formic acid. Further elution with acetonitrile-formic acid mix (35μL of 50% acetonitrile, 0.2% formic acid) helped maximize recovery of hydrophobic peptides. Finally, peptides were dried in a speed-vac and resuspended in buffer MS2 [4% acetonitrile, 0.1% formic acid] for subsequent mass spectrometry analysis.
Peptides (~1.5μg) were injected into an Ultimate 3000 UHPLC system (Thermo Scientific) comprising a PepMap100 C18 (75μm ID x 2cm, particle size 3μm, pore size 100A; Thermo Fisher Scientific) precolumn upstream of a PepMap C18 (75μm ID x 2cm, particle size 3μm, pore size 100A; Thermo Fisher Scientific) analytical column. Sample loading proceeded at 300 nL/min in 98% buffer LCMS_A [0.1% formic acid in water], 2% LCMS_B [0.1% formic acid in acetonitrile]. After washing the precolumn with 20 column volumes of LCMS_A, peptides were resolved using a 120-min linear gradient from 4% to 30% LCMS_B. Peptides were ionized by nanospray electrospray ionization and analyzed on a Q Exactive HF-X mass spectrometer (Thermo Scientific). For data-dependent acquisitions, MS1 full scans (350-1400 m/z range) were collected at a resolution of 60,000 (AGC 3x106, 50ms maximum injection time). HCD fragmentation of the 12 most abundant precursor ions was performed at 25% NCE. The fragment analysis (MS2) was performed with a resolution of 15,000 (AGC 1x105, 100ms maximum injection time, 2.2 m/z isolation window). For data-independent acquisitions, MS1 full scans (350-1400 m/z range) were collected at a resolution of 120,000 (AGC 3x106, 50ms maximum injection time). Fragment analysis (MS2) was subdivided into 25 DIA isolation windows of varying widths (see Supplementary Table 1) using a resolution of 30,000 (AGC 1x106, 70ms maximum injection time). HCD fragmentation was performed at 25% NCE. The resulting raw DIA files were analyzed using Spectronaut (v15), with a search database containing the E. coli Uniprot database concatenated to a database containing common contaminants. In silico digests were performed using Trypsin/P and LysC. The false discovery rate was set at 1% at both the peptide and protein levels. Normalization of all replicates and generation of the volcano plot were performed using Spectronaut.
For the preparation of samples for mass spectrometric analysis of the pull-down of FtsH from wild-type and ΔhflK/C strains, the same purification protocol for the FtsH•HflK/C complex describe above was followed, using GDN detergent to retain weak interactions between FtsH and its substrates. After running an SDS-PAGE gel of the FLAG resin eluate from WT and ΔhflK/C cells, the entire gel lane for each condition was excised and sent to the proteomic core facility at the Koch Institute for Cancer Research (MIT) for data analysis. There, samples were analyzed via DDA on a Q Exactive HF-X mass spectrometer coupled to a Ultimate 3000 UHPLC. Co-purifying proteins in each condition were quantified using spectral counts, normalized to that of FtsH in each sample.
Biochemical assays.
Expression and purification of FtsH-bearing samples followed protocols listed above, using the following buffer substitutions. Buffer A was substituted with buffer BA1 [50mM Tris-HCl (pH 8.0), 1mM MgCl2, 100mM KCl, 10% glycerol, and 3mM β-mercaptoethanol] during cell lysis, and with buffer BA2 [50mM Tris-HCl (pH 8.0), 100mM NaCl, 5mM MgCl2, 100μM ZnCl2, 10% glycerol, 3mM β-mercaptoethanol] when resuspending the pelleted membranes. Additionally, prior to elution, the resin was washed using buffer BA3 [50mM Tris-HCl (pH 8.0), 400mM NaCl, 5mM MgCl2, 100μM ZnCl2, 0.03% DDM and 10% glycerol]. The protein was eluted with 0.25 mg/mL FLAG peptide (APEXBio) in buffer BA5 [50mM Tris-HCl (pH 8.0), 100mM NaCl, 5mM MgCl2, 100μM ZnCl2, 10% glycerol, and 0.03% DDM] according to the manufacturer’s directions.
The λ-CII substrate was produced using HisTag-SUMO-λ-CII in the pET22b plasmid. The BL21(DE3) cells harboring the plasmid were grown to an OD600 of ~0.8 at 37°C. Protein expression was induced by addition of 0.5mM of IPTG, and the culture was grown overnight at 18°C. Cells were harvested by centrifuging at 4,000rpm for 25 min at 4°C. The cell pellet was resuspended in buffer BA6 [20mM HEPES (pH 8.0), 400mM NaCl, 100mM KCl, and 10% glycerol], and stored at −80°C.
For purification, the cells were thawed and incubated with 1 mM DTT and protease inhibitor cocktail prior to lysis using 30% power with cycles of 10 s on and 30 s off for a total duration of 3.5 min. The sample was centrifuged at 4,500 rpm for 30 min in a JA25.50 rotor to remove cell debris. The supernatant was transferred to a new tube and incubated with pre-washed Ni-NTA resin for 30 minutes at 4°C in buffer BA6 supplemented with 5mM imidazole. The resin was washed with 200 mL of buffer BA6 supplemented with 15mM imidazole to remove impurities. The resin was then incubated with the Sumo Protease overnight at 4°C in buffer BA6 containing 5mM imidazole. The next day, the cleaved protein was collected as HisTag-SUMO remained bound to the resin. The cleaved protein was concentrated using a 10-kDa cutoff centrifugal unit and loaded onto a S200 (16/600) gel filtration column. Fractions were collected, analyzed using SDS-PAGE, concentrated, aliquoted, and flash-frozen in liquid nitrogen for future use.
Degradation assays were performed at 37 °C using 5μM λ-CII, ~0.3μM of FtsH•HflK/C complex in buffer BA7 [25mM HEPES (pH 7.5), 150mM NaCl, 5mM MgCl2, 3mM β-mercaptoethanol,100μM ZnCl2, 5mM ATP, 0.009% DDM]. The concentrations of FtsH/HflK/C were calculated using a BSA standard after running SDS PAGE. All components except β-CII were initially mixed at 37°C for 5 min. λ-CII, which had also been preincubated at 37°C, was then added to initiate degradation, which was monitored by SDS-PAGE gel analysis.
ATP hydrolysis was measured using a coupled enzymatic reaction (Norby, 1988) in which NADH oxidation to NAD+ reduces absorbance at 340 nm (Δε = 6.22 mM−1cm−1) using a SpectraMax M5 plate reader and a 384-well assay plate (Corning, 3575). A 20X stock ATPase reaction mix contained 20μL of pyruvate kinase and lactic dehydrogenase from rabbit muscle (P0294, Sigma Aldrich), 10μL of 200mM NADH, grade II (CAS# 606688), 15μL of 1M phosphoenolpyruvate (Sigma Aldrich, 10108294001) in 25mM HEPES-KOH (pH 7.6), and 25μL of 200mM ATP. For assays, FtsH6 (0.5 μM) was present in 10μL of buffer BA7. After 5 min of equilibration at 37°C, the ATPase assay was initiated by addition of an equal volume of 1X ATPase reaction mix.
Lipid preparation.
All lipids, including POPC (#850457C), POPE (#850757C), and NBD-PE (#810153C), were purchased from Avanti lipids, and liposomes were prepared as described (Ghanbarpour et al., 2021, Ploier and Menton, 2016, Marek and Gunther-Pomorski, 2016)). Briefly, a mixture of POPC (90% of total lipid by weight), POPE (9.5%), and NBD-lipid (0.5%, either NBD-PE or NBD-PC) solubilized in chloroform was mixed and dried under a nitrogen stream followed by vacuum to form a lipid film. The dry lipid film was subsequently resuspended in buffer LP1 [50mM HEPES (pH 7.6), 200mM NaCl] to create a 10.5mM lipid stock, which was incubated at 37°C for 10 min, followed by resuspension and ten freeze-thaw cycles. Finally, the lipid mixture underwent 30 cycles of extrusion through a 400-nm polycarbonate filter to complete the preparation.
Proteoliposome preparation.
Liposomes, with a lipid concentration of 5.25mM in a total volume of 250μL, were subjected to destabilization by adding Triton X-100. The concentration of Triton X-100 used was determined based on a prior swelling assay (Ghanbarpour et al. 2021). This destabilization process was carried out at room temperature for 2-3 h. Proteins solubilized in detergent were then introduced to the destabilized liposomes, and the mixture was rotated gently for 1 h to ensure mixing. Removal of detergent was achieved in three stages using pre-washed biobeads (Bio-Rad #152-3920). Initially, 20mg of biobeads were added to the mixture, which was then incubated at room temperature for 1 h. Subsequently, 20mg of additional biobeads were added, and the mixture was allowed to rotate at room temperature for 2 h. In the final step, the mixture was transferred into a new tube containing 40mg of fresh biobeads and rotated at 4°C overnight to complete detergent removal, prior to removing biobeads by pipetting.
Scramblase assay.
Lipid scramblase activity was monitored at 30°C using 96-well plates containing 100 μL of either liposomes or proteoliposomes. The assay tracked the decrease in NBD fluorescence (excitation 460 nm; emission 538 nm) upon the introduction of dithionite to a final concentration of 5mM using a BioTek Synergy H1 Hybrid Multi-Mode Reader. Subsequently, an additional dose of dithionite (5mM) along with Triton X-100 (0.5%) was added to dissolve liposomes and ensure NBD reduction.
To test for proteolipid disruption, a 1-myristoyl-2-C6-NBD-PE bovine serum albumin (BSA) back-extraction assay (Ghanbarpour et al., 2021; Marek and Gunther-Pomorski, 2016; Ploier and Menon, 2016) was carried out adding 3mg/mL BSA to liposomes or proteoliposomes instead of dithionite. Like dithionite, BSA interacts exclusively with NBD-PE from the outer leaflet of liposomes, but BSA quenches fluorescence less effectively than dithionite. The protocol used above was used for the NBD-glucose assay, except NBD-lipids were excluded from liposomes or proteoliposomes and NBD-glucose at a concentration of 12.6 μM was introduced during the liposome-destabilization phase. Glucose leakage was assessed after 2 days of dialysis against buffer free of NBD-glucose.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01-GM144542 and R35-GM141517, and NSF-CAREER grant 2046778. Samples were prepared at the Automated Cryogenic Electron Microscopy Facility in MIT.nano and screened on a Talos Arctica microscope, which was a gift from the Arnold and Mabel Beckman Foundation. We thank Dr. Andrew V. Grassetti for the gift of graphene-coated grids.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
DATA AND SOFTWARE AVAILABILITY
The correspondence for material and data in the manuscript should be addressed to jhdavis@mit.edu or alirezag@wustl.edu The raw electron micrographs and mass spectrometry data will be publicly deposited upon publication. Density maps and atomic models are available through the EMDB and the PDB.
REFERENCES
- Amberg-Johnson K, Hari SB, Ganesan SM, Lorenzi HA, Sauer RT, Niles JC, Yeh E (2017) Small molecule inhibition of apicomplexan FtsH1 disrupts plastid biogenesis in human pathogens. eLife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends J, Thomanek N, Kuhlmann K, Marcus K, Narberhaus F (2016) In vivo trapping of FtsH substrates by label-free quantitative proteomics. Proteomics 16: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT (2012) ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1823: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner LM, Arends J, Narberhaus F (2017) When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol Chem 398: 625–635 [DOI] [PubMed] [Google Scholar]
- Browman DT, Hoegg MB, Robbins SM (2007) The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol 17: 394–402 [DOI] [PubMed] [Google Scholar]
- Casanal A, Lohkamp B, Emsley P (2020) Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci 29: 1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Ito K, Akiyama Y (2006) The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol Microbiol 60: 448–457 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux PF, Herrmann A, Ohlwein N, Kozlov MM (2008) How lipid flippases can modulate membrane structure. Biochimica et Biophysica Acta (BBA) - Biomembranes 1778: 1591–1600 [DOI] [PubMed] [Google Scholar]
- Fu Z, MacKinnon R (2024) Structure of the Flotillin Complex in a Native Membrane Environment. bioRxiv: 2024.2005.2009.593390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbarpour A, Valverde DP, Melia TJ, Reinisch KM (2021) A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc Natl Acad Sci U S A 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glueck D, Grethen A, Das M, Mmeka OP, Patallo EP, Meister A, Rajender R, Kins S, Raschle M, Victor J et al. (2022) Electroneutral Polymer Nanodiscs Enable Interference-Free Probing of Membrane Proteins in a Lipid-Bilayer Environment. Small 18: e2202492. [DOI] [PubMed] [Google Scholar]
- Grassetti AV, May MB, Davis JH (2023) Application of Monolayer Graphene to Cryo-Electron Microscopy Grids for High-resolution Structure Determination. J Vis Exp [DOI] [PubMed] [Google Scholar]
- Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D’Ari R, Bouloc P (1993) Cell growth and lambda phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci U S A 90: 10861–10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA (2003) Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell 11: 659–669 [DOI] [PubMed] [Google Scholar]
- Hinz A, Lee S, Jacoby K, Manoil C (2011) Membrane proteases and aminoglycoside antibiotic resistance. Journal of Bacteriology 193: 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Akiyama Y (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Jarsch IK, Daste F, Gallop JL (2016) Membrane curvature in cell biology: An integration of molecular mechanisms. Journal of Cell Biology 214: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Akiyama Y, Ito K (1996) A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J 15: 6122–6131 [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Akiyama Y, Ito K (1998) Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA11Edited by Karn J.. Journal of Molecular Biology 279: 175–188 [DOI] [PubMed] [Google Scholar]
- Kinman LF, Powell BM, Zhong ED, Berger B, Davis JH (2023) Uncovering structural ensembles from single-particle cryo-EM data using cryoDRGN. Nat Protoc 18: 319–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langklotz S, Baumann U, Narberhaus F (2012) Structure and function of the bacterial AAA protease FtsH. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1823: 40–48 [DOI] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ et al. (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loser T, Joppe A, Hamann A, Osiewacz HD (2021) Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging. Cells 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Wang C, Luo D, Yan L, Yang W, Li N, Gao N (2022) Structural insights into the membrane microdomain organization by SPFH family proteins. Cell Research 32: 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek M, Gunther-Pomorski T (2016) Assay of Flippase Activity in Proteoliposomes Using Fluorescent Lipid Derivatives. Methods Mol Biol 1377: 181–191 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, Held SR (2017) Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol 197: 102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norby JG (1988) Coupled assay of Na+,K+-ATPase activity. Methods Enzymol 156: 116–119 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30: 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploier B, Menon AK (2016) A Fluorescence-based Assay of Phospholipid Scramblase Activity. J Vis Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BM, Brant TS, Davis JH, Mosalaganti S (2024) Rapid structural analysis of bacterial ribosomes in situ. bioRxiv: 2024.2003.2022.586148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BM, Davis JH (2024) Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN. Nature Methods [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Yokoyama T, Yan X-F, Beh IT, Shi J, Basak S, Akiyama Y, Gao Y-G (2022) Cryo-EM structure of the entire FtsH-HflKC AAA protease complex. Cell Reports 39 [DOI] [PubMed] [Google Scholar]
- Rei Liao JY, van Wijk KJ (2019) Discovery of AAA+ Protease Substrates through Trapping Approaches. Trends Biochem Sci 44: 528–545 [DOI] [PubMed] [Google Scholar]
- Reisch CR, Prather KLJ (2017) Scarless Cas9 Assisted Recombineering (no-SCAR) in Escherichia coli, an Easy-to-Use System for Genome Editing. Curr Protoc Mol Biol 117: 31.38.31–31.38.20 [DOI] [PubMed] [Google Scholar]
- Sadeghi S, Müller M, Vink RL (2014) Raft formation in lipid bilayers coupled to curvature. Biophys J 107: 1591–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikawa N, Akiyama Y, Ito K (2004) FtsH exists as an exceptionally large complex containing HflKC in the plasma membrane of Escherichia coli. J Struct Biol 146: 123–129 [DOI] [PubMed] [Google Scholar]
- Singh S, Darwin AJ (2011) FtsH-dependent degradation of phage shock protein C in Yersinia enterocolitica and Escherichia coli. J Bacteriol 193: 6436–6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kinman LF, Jahagirdar D, Ortega J, Davis JH (2023) KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion. Nat Struct Mol Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, Lyumkis D (2017) Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods 14: 793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D, Cramer P (2019) Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods 16: 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D, Xue L, Dienemann C, Cramer P, Mahamid J (2021) Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 A in cells. Nat Methods 18: 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman AJ, Oppenheim AB, Yura T, Yamanaka K, Niki H et al. (1995) Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. Embo j 14: 2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas JL, Gomez-Blanco J, Conesa P, Melero R, Miguel de la Rosa-Trevin J, Oton J, Cuenca J, Marabini R, Carazo JM, Vargas J et al. (2018) MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure 26: 337–344 e334 [DOI] [PubMed] [Google Scholar]
- Wessel AK, Yoshii Y, Reder A, Boudjemaa R, Szczesna M, Betton JM, Bernal-Bayard J, Beloin C, Lopez D, Völker U et al. (2023) Escherichia coli SPFH Membrane Microdomain Proteins HflKC Contribute to Aminoglycoside and Oxidative Stress Tolerance. Microbiol Spectr 11:e0176723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal K, Langklotz S, Thomanek N, Narberhaus F (2012) A trapping approach reveals novel substrates and physiological functions of the essential protease FtsH in Escherichia coli. J Biol Chem 287: 42962–42971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G, Rapoport TA (2020) Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Liu B, Nixon PJ, Yu J, Chen F (2022) Recent Advances in Understanding the Structural and Functional Evolution of FtsH Proteases. Frontiers in Plant Science 13: 837528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong ED, Bepler T, Berger B, Davis JH (2021) CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods 18: 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The correspondence for material and data in the manuscript should be addressed to jhdavis@mit.edu or alirezag@wustl.edu The raw electron micrographs and mass spectrometry data will be publicly deposited upon publication. Density maps and atomic models are available through the EMDB and the PDB.