Abstract
The ubiquitous RNA-processing molecule TDP-43 is involved in neuromuscular diseases such as inclusion body myositis, a late-onset acquired inflammatory myopathy. TDP-43 solubility and function are disrupted in certain viral infections. Certain viruses, high viremia, co-infections, reactivation of latent viruses, and post-acute expansion of cytotoxic T cells may all contribute to inclusion body myositis, mainly in an age-shaped immune landscape. The virally induced senescent, interferon gamma-producing cytotoxic CD8+ T cells with increased inflammatory, and cytotoxic features are involved in the occurrence of inclusion body myositis in most such cases, in a genetically predisposed host. We discuss the putative mechanisms linking inclusion body myositis, TDP-43, and viral infections untangling the links between viruses, interferon, and neuromuscular degeneration could shed a light on the pathogenesis of the inclusion body myositis and other TDP-43-related neuromuscular diseases, with possible therapeutic implications.
Keywords: TDP-43, Inclusion body myositis, Myositis triggers, Interferon gamma, Long COVID
Background
Inclusion body myositis (IBM) is an inflammatory myopathy occurring after middle age, with autoimmune and degenerative mechanisms [1, 2]. Other idiopathic inflammatory myopathies (IIMs) are dermatomyositis (DM), polymyositis (PM), overlap syndromes including anti-synthetase syndrome and necrotizing pauci-immune myositis [3]. The distinction between IBM, PM, and PM with mitochondrial pathology is not neat, raising the question whether IBM is a variant of PM occurring in the older age, related to immunosenescence [4]. IBM pathogenesis centrally involves cytotoxic, senescent CD8+ T cells, defects of autophagy and ubiquitin–proteasome system (UPS) resulting in proteostasis impairment and abnormal sarcoplasmic protein aggregation, along with endoplasmic reticulum and mitochondrial alterations, and antibodies to the cytosolic 5′-nucleotidase 1A (anti-cN1A) [1, 5]. The driving mechanisms of this pathology, however, are still evasive.
IBM belongs to a group of neurological disorders, the TDP-43 proteinopathies, which pathogenically involve TDP-43 [TAR-DNA-binding protein 43 (transactive response DNA-binding protein of 43 kDa)] [6]. TDP-43, encoded by the TARDBP gene, an RNA- and DNA-binding nuclear regulatory protein, member of the heterogeneous nuclear ribonucleoprotein (hnRNP) family [7, 8]. In skeletal muscles, TDP-43 is involved in transcription regulation, RNA splicing, mRNA stability, RNA transport, and quality control and undergoes post-translational modifications with functional consequences [9]. TDP-43 functions in muscles are complex, including myoregeneration (Table 1). In neurodegeneration, the mechanisms of TDP-43 involvement include cytotoxic aggregations, nuclear loss, alteration of cellular functions, and others [6, 10].
Table 1.
Roles of TDP-43 in muscles
| Function | Roles of TDP-43 | References |
|---|---|---|
| mRNA metabolism |
TDP-43 is involved in transcription regulation, nucleocytoplasmic shuttling, mRNA splicing, translation, transcription, transport, stabilization, miRNA, and lncRNA processing, and RNA quality control TDP-43 binds ssRNA and DNA and acts as a transcription repressor, or scaffold for nuclear bodies |
[1–3, 9, 11–14] |
| Myogenesis |
TDP-43 is involved in muscle development and differentiation, neuromuscular junction formation, and muscle regeneration after injury TDP-43 transiently forms during myogenesis amyloid-like myogranules, along with RNA- and RNA-binding proteins TDP-43 is required for the expression of myogenesis regulators and myogenic microRNAs such as miR-1 and miR-206 [9, 13] TDP-43 activates Wnt/β-catenin signaling, involved in muscle regeneration and fibrosis |
[5, 6, 9, 13, 15–18] |
|
Association with mitochondria |
In myogranules, TDP-43 co-localizes and interacts with the mitochondrial inner membrane protein CHCHD10 . In IBM, TDP-43 aggregates accumulate with mitochondria in myofibers, resulting in mitochondrial and muscle fibers toxicity. TDP-43 targets the mitochondria complex I |
[6, 19, 20] |
IFN interferon, RBP ribonucleoproteins, lncRNA long non-coding RNAs, ssRNA single-strand RNA
IBM muscle biopsies reveal cytoplasmic aggregation of TDP-43 and TDP-43 nuclear loss [10]. Even an 1% amount of myofibers staining for TDP-43 in a muscle biopsy was highly sensitive and specific for IBM [11].
TDP-43 may have an emerging intriguing role in viral infections [12]. TDP-43 is involved in controlling IFN responses triggered by endogenous RNA, but the TDP-43 role as an RNA-binding protein in viral infections is rarely investigated [13, 14]. Loss of TDP-43 results in dsRNA intracellular accumulation and interferon (IFN) triggering [13]. The TDP-43 ortholog of Caenorhabditis elegans called TDP1 limits dsRNA accumulation [21]. Also, knockdown of TARDBP increases viral replication in macrophages [14] and TDP-43 knockdown amplifies enterovirus infections, suggesting an antiviral effect of TDP-43 [22]. Moreover, TDP-43 binding is protective against HIV-1 by sterically hindering a HIV-1 promoter [23]. Also, after TDP-43 knockdown in mouse brain, the type I IFN-inducible genes, including the mouse orthologs of the intracellular sensor molecules RIG-I and MDA-5 which detect viral RNA, are the most overexpressed [21, 24]. In Coxsackie B3 infection, the viral protease 2A alters TDP-43 distribution, solubility, and function [22]. Therefore, TDP-43 could have an important role in the viral-induced IFN response in TDP-43 proteinopathies, including IBM (Table 2).
Table 2.
Immunomodulatory and antiviral roles of TDP-43
| Function | Roles of TDP-43 | References |
|---|---|---|
| Immunomodulatory |
TDP-43 regulates the accumulation of RNA polymerase III transcripts and other endogenous immunostimulatory dsRNAs which trigger IFN TDP43 limits overexpression of IFN-I related genes including RIG-I and MDA-5 in animal models TDP-43 interacts with lncRNAs such as Malat1, which prevents TDP-43 cleavage and IFN generation TDP-43 aggregation may be induced by IFN-ƴ and by low amounts of cytoplasmic RNA TDP-43 expression activates GSK3, which delays and decreases IFN-1 production and enhances IFNγ and other pro-inflammatory cytokines production GSK is involved in TDP-43 phosphorylation and aggregation |
[9, 11, 25–34] |
| Antiviral | TDP-43 binds YB-1, a host regulator of HCV replication | [35] |
|
TDP-43 suppresses HIV1 transcription by binding HIV-1 long terminal repeats HIV-1 could replicate in human immune cells independent of TDP-43 A specific deubiquitinase inhibitor, IU1, reversed HIV-1 latency by degrading TDP-43 Knocking down TDP-43 with siRNAs in cell cultures reactivates HIV-1 by reversing its latency TDP-43 binding may sterically hinder the HIV-1 LTR promoter involved in viral transcription and reactivation Silencing TDP-43 increases HIV-1 infectivity by reducing HDAC6 |
[23] |
|
| TDP-43 is protective against enteroviruses | [38] | |
| TDP-43 RRM binds the SARS-CoV2 S1 RBD | [39] |
dsRNA double-strand RNA, IFN interferon, RBP ribonucleoproteins, lncRNA long non-coding RNAs, Malat1 metastasis-associated lung adenocarcinoma transcript-1, GSK3 glycogen synthase kinase 3, RRM RNA recognition motif 1, SARS-CoV2 S1 RBD SARS-CoV-2 spike S1 protein receptor binding domain, RBPs RNA-binding proteins, YB-1-box-binding protein-1
Role of TDP-43
TDP-43 in basal conditions and in infections
In basal conditions, TDP-43 is bound by the long-non-coding RNA (lncRNA) Malat1 (metastasis-associated lung adenocarcinoma transcript-1), in humans called MALAT1 [14]. Malat1 binding hinders the TDP-43 cleavage, mediated by activated caspase-3, from generating TDP-35 and IRF3 (IFN regulator factor 3) [14]. Generally, viral infections result in reduced expression of Malat1, promoting antiviral IFN production [14]. Moreover, TDP-35 amplifies the IFN–I production by degrading the negative regulator of IRF3 called Rbck1 (RanBP-type and C3HC4-type zinc finger-containing protein 1) [14]. However, Malat1 function may increase in certain viral infections, such as HIV, Coxsackie myocarditis or mild COVID-19 [14, 25, 40]. Malat1 has immunosuppressor and NF-kB- and NLRP3 regulatory effects [40, 41]. Also, MALAT1 is upregulated in IBM [42]. Increased Malat1 in IBM and TDP-43 aggregation may likely depend on viral characteristics and is in line with a slow inflammatory response.
TDP-43 can be activated after caspase-induced cleavage, the N-terminal cleavage product of TDP-43 forming protein aggregates, while the C-terminal cleavage product is degraded by proteasomes [22]. The proteasome inhibition contributes to the pathogenesis of IBM, as the major proteasomal enzymes have decreased activity [43]. Immunoproteasomes (iPS) found in immune tissues (constitutively expressed in hematopoietic cells or induced in response to IFN gamma or TNF alpha) have structural similitudes to proteasomes but have three different inducible catalytic subunits (PSMB8, -B9, and -B10), triggered by IFN-ƴ in viral infections, or by other pathogens, proteins, or particles [44–46]. Generation of cytotoxic CD8+ T cell responses upon a viral infection requires antigen processing through the proteasome, which selectively cleaves after certain ammino acids residues [46].
TDP-43 in IBM biopsies
In IBM muscle biopsies, the overexpression of the immunoproteasome (iPS) subunits PSMB8 and-9, correlated with IFN-ƴ, IRF1 (interferon regulatory factor 1), and STAT1 (signal transducer and activator of transcription 1), is another argument for a viral trigger [44, 47]. The iPS upregulates the major histocompatibility complex MHC-1 and MHC-2 on myofibers, exposing them to immune attack [48–50]. Also, iPS are involved in muscle remodeling and prevention of protein aggregation [51]. Of note, mutations of the (immuno)proteasome subunits, as in the rare autoinflammatory diseases Nakajo–Nishimura or CANDLE syndrome, may result in an IBM-like myositis [52].
IBM patients have a high IFN score and IFN-ƴ signature, along with increased IFN type I expression in muscle which amplifies inflammation [53, 54]. The IFN-ƴ, central in IBM, is produced by the highly differentiated cytotoxic CD8+ T cells, reprogrammed with aging to fulfill innate-like functions [55, 56]. These CD8+ cells and effector memory T cells re-expressing CD45RA (TEMRA) found in IBM may be induced not only by senescence, but also by persistent viruses [1, 4, 55, 57]. IFN-ƴ induces ER stress and aggregation of TDP-43 and other proteins [1, 5, 54]. IFN-I may also be induced by anti-Ro52, present in some IBM patients [50, 58]. Ro52 or TRIM21 (tripartite motif proteins), is an IFN–inducible E3 ligase involved in IFN type I downregulation [59]. Other infection-related factors may intervene in IBM, such as activation of NLRP3 inflammasome, heat shock proteins (HSP), ribosomal proteins, or molecular mimicry with a mycobacterial protein guanylate-binding protein 2 (GBP2) with antiviral and anti-tuberculous functions [5, 60]. GBP2 is involved in the control of mRNA splicing [5] with possible relevance in TDP-43 dysfunction when mRNA splicing is altered.
Also, the glycogen synthase kinase 3 (GSK3), a serine/threonine kinase with 2 isoforms (α and β), is activated in IBM [33]. GSK3, involved in many cellular processes, is an immunomodulator in IBM [33]. GSK3 delays and decreases IFN-1 production, enhances IFNγ signaling, but also increases and delays pro-inflammatory cytokines production [33]. Moreover, GSK3β is one of the protein kinases involved in the TDP-43 phosphorylation [34]. TDP-43 expression activates GSK3, and GSK inhibition decreases TDP-43 aggregation [35].
Also, activation of autophagy is part of the innate immune response, and autophagy receptors may become viral targets [15]. Amongst these autophagy receptors, NBR1 (neighbor of BRCA1), a ubiquitin-binding scaffold protein, increases in viral infections [61], and NBR1 accumulates and is abnormal in IBM muscle [62].
In IBM, the dysregulation of a deubiquitinase called cylindromatosis (CYLD) reduces the autophagic clearance of protein aggregates [63]. CYLD is expressed with phosphorylated TDP-43 in the sIBM myofibers [63]. CYLD, required for antiviral host defense, is involved in the STING cleavage [64] and negatively regulates NF-kB [63].
IFN-ƴ and low RNA amounts in cytoplasm also stimulate aggregation of TDP-43 and other RBPs with “prion-like” low-complexity (LC) domains, favored by proteins misfolding in aging [1, 65].
Potential links between IBM and viral infections
The IBM occurrence may reflect various pathogenic associations, including viral infections [4]. In general, chronic IIM may be triggered by viruses such as Coxsackie B, enterovirus, parvovirus, HTLV-1, or HIV [66]. Mechanisms of viral-induced myositis hypothetically include direct invasion of myocytes by the virus, molecular mimicry, exposure of cryptic epitopes after conformational alterations, myotoxic cytokines such as IFNs and autoimmune reactions [66–68]. Latent viral infection, viral-induced denaturation of self-structures or homologies with various viral proteins could result in a prolonged immune response [66]. For instance, enterovirus 71 (EV71) may upregulate TRIM21 (Ro52), which degrades SAMHD1, a host antiviral molecule [59]. Also, during a viral infection, many ribonucleoproteins including TDP-43, are hijacked [12]. Coxsackie virus B3 protease 3C causes TDP-43 cytoplasmic redistribution and aggregation [12, 22].
Also, the aging cellular environment may make the myofiber susceptible to a newly invading virus, or may allow cytopathic manifestation of a virus, or a vertically transmitted genomic endogenous virus such as a retrovirus dormant for years, such as HTLV1, may start to be transcribed due to the age-modified milieu [16]. Endogenous retroviruses (ERVs, genomic remnants of ancient viral infections, most inactive and non-infectious) are mutually reinforcing with TDP-43 proteinopathies regarding neurodegeneration [17, 26]. Moreover, aging may favor both ERVs expression and TDP-43 proteinopathy [26].
However, no definite evidence for a viral etiology of IBM has been established [27]. Mumps virus was described as a potential IBM cause, later questioned in immunohistochemical studies [28]. IBM patients have an increased prevalence of hepatitis C virus (HCV) or human lymphotropic T virus-1 (HTLV1) [69–71]. The relationship between HCV and TDP-43 is yet to be clarified. TDP-43 binds YB (Y-box-binding protein-1), a host factor involved in HCV capsids assembling, and TDP-43 knockdown significantly decreased HCV replication [19]. The persistent HCV-related IFN upregulation and lymphocyte exhaustion may in fact contribute to the chronic myopathy in HCV [4]. TDP-43 facilitates HBV gene expression stimulating its transcription and assembly of protein complexes [12]. Furthermore, the clinical picture of IBM patients with HCV is different from the one of patients with IBM and HIV; therefore, no unique mechanism links a chronic viral infection to IBM [20].
Most of the HIV-positive patients with myositis had overlapping features of PM and IBM, which clinically progress to IBM, and most of them have anti-c1NA antibodies and rimmed vacuoles [20]. TDP-43 suppresses HIV-1 transcription by binding HIV-1 long terminal repeat [72]. Knocking down TDP-43 with siRNAs in cell cultures reactivates HIV-1 by reversing its latency [23]. Notwithstanding, HIV-1 can replicate in human immune cells independent of TDP-43 [73]. In viral-associated IBM in HIV and HTLV-1, the viral antigen is not present in myofibers but in the T cells and macrophages instead [1]. HIV infection can induce T cells immune senescence [74]. Thus, it is more conceivable that the virally induced senescent, IFN-ƴ producing cytotoxic CD8+ T cells lead to IBM.
IBM has been reported to be induced by Covid-19 in a 54-year female patient with diabetes mellitus and hyperlipidemia on statins [75]. Also, an axial paraspinal myopathy was reported in Covid-19 [76], and paraspinal myositis may be a feature of IBM [27]. However, long-term consequences of SARS-CoV2 infection, including muscular involvement, are starting to be recognized [77]. After COVID-19, the prevalence of myositis-specific antibodies and myositis-associated antibodies increases [78]. Possible mechanisms include type I IFN pathways, NLRP3 inflammasome activation, or a previous exposure to common coronaviruses [79]. SARS-CoV-2 impairs the stress granules (SGs) disassembly, and the SARS CoV-2 nucleocapsid N protein binds the SG-related amyloid proteins, favoring aggregation [24]. Also, SARS-CoV-2 spike S1 protein receptor binding domain (SARS-CoV2 S1 RBD) attaches to TDP-43 RRM at the viral surface, initiating aggregation [39]. TDP-43 is aggregated and hyperphosphorylated in SARS-CoV2 patients [12]. Also, the SARS-COV2 nucleocapsid N protein phosphorylation is mediated by GSK3, delaying the IFN-1 response [33] (Fig. 1).
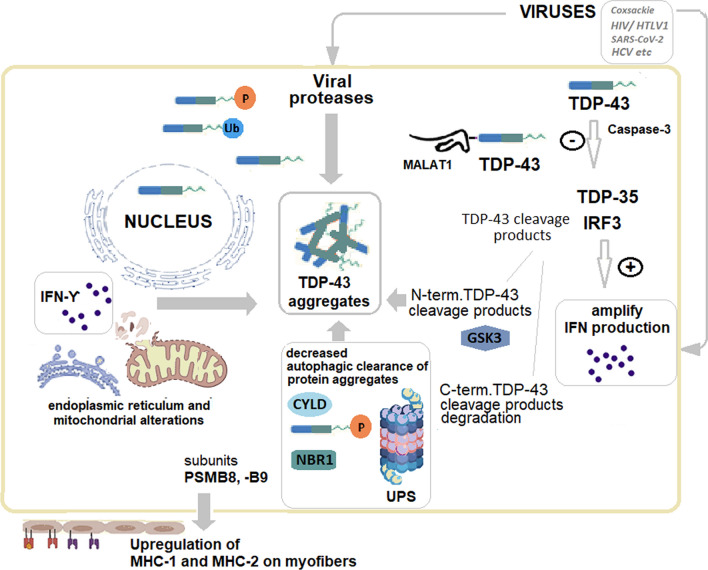
Fig. 1.
Regulation of TDP-43 in viral infections: potential implications for cellular processes in IBM pathogenesis
Legend: In IBM, TDP-43 becomes mislocalized and accumulates in the cytoplasm of cells, leading to protein aggregation and disruption of many cellular processes. Into the myofibers, in basal conditions, TDP-43 is bound by the long-non-coding RNA (lncRNA) Malat1 (metastasis-associated lung adenocarcinoma transcript-1). Malat1 binding prevents the TDP-43 cleavage, mediated by activated caspase-3, to generate TDP-35 and IRF3 (IFN regulator factor 3). TDP-35 amplifies the IFN production by degrading the negative regulator of IRF3. Generally, viral infections result in reduced expression of Malat1, promoting antiviral IFN production. However, certain viruses (Coxsackie B, hepatitis C, HIV, HTLV-1, SARS-CoV2, etc.) increase Malat1, delaying and decreasing IFN-1 production. Nevertheless, the implications for IBM pathogenesis are still hypothetical. GSK3 (glycogen synthase kinase 3) similarly enhances IFNγ and pro-inflammatory cytokines production, phosphorylating TDP-43 and promoting TDP-43 aggregation. After caspase-induced cleavage, the N-terminal cleavage product of TDP-43 may form protein aggregates, while the C-terminal cleavage product is degraded by proteasomes. Autophagy receptors may become viral targets. Amongst these autophagy receptors, NBR1 (neighbor of BRCA1), a ubiquitin-binding scaffold protein, increases in viral infections, and NBR1 accumulates and is abnormal in IBM muscle. Defects of autophagy and ubiquitin–proteasome system (UPS) result in proteostasis impairment and abnormal sarcoplasmic protein aggregation. TDP-43 is involved in the viral-induced IFN response, inducing mitochondrial and endoplasmic reticulum damage, and activating mitochondrial unfolded protein response. IFN gamma plays a major role in these processes.
Muscle weakness or fatigue frequently persists over 6 months after SARS-CoV2 infection, accompanied by electrophysiological myopathic changes [29, 80]. Post-acute COVID-19 sequelae (PASC) may affect 1/3 up to 2/3 of COVID-19 patients [30, 81]. PASC may be shaped by factors like endothelial damage, immunosenescence, mitochondrial alterations, and possibly by higher viral burden, and others [82]. In a longitudinal multi-omics study, SARS-CoV2 viremia, reactivation of latent viruses such as cytomegalovirus (CVM) and Epstein-Barr virus (EBV) and post-acute expansion of cytotoxic T cells were amongst the factors associated with PASC [81]. A particular PASC proinflammatory immune endotype, enriched with cytotoxic effector signatures in CD8+ and NK, has been identified [81]. It is tempting to speculate that co-infection with other viruses such as CMV could “flatten” the IFN-α initial production and stimulate persistent CD8+ T cells with IFN-ƴ production in long-Covid.
Moreover, not only cytotoxic CD8+ T cells, but also the plasma cell infiltrate from the muscles of IBM patients has a distinct B cell receptor repertoire, different from DM and PM, reflecting features of antigen-driven selection and differentiation [83]. It could be speculated that the T cells and plasma cell expansions may reflect linked recognition of common antigens, which needs further study in IBM [1, 83]. Moreover, in some IBM patients, there is an increased population of large granular T lymphocytes (T-LGL) characterized by augmented expression of surface molecules KLRG1 and CD57 [1]. The expanded T-LGL in IBM are rather secondary, “reactive,” with a senescent-like profile, associated with inhibitory NK cell receptors and increased inflammatory and cytotoxic features [84]. Of interest, HIV-1 infection is also a risk factor for the evolution of clonal T-LGL disorders [85].
In IBM as in other autoimmune diseases, immunoaging may come with an increased risk for autoimmunity, possibly the price to pay to preserve some of the immune competency [86]. The virally induced senescent, IFN-ƴ producing cytotoxic CD8+ T cells may be the ones involved in IBM, in a predisposed host.
Genetics in IBM and viral infections
Susceptibility genes for IBM include HLA DRB1*03:01, 01:01, and 13:01 alleles, respectively [87–89]. The HLA-DRB1*03 allele, as a component of the ancestral HLA 8.1 haplotype, is a susceptibility factor for IIMs and many other autoimmune diseases [90]. An arginine in position 74 of the DRβ1 chain confers the allelic risk for IBM [89]. HLA DRB1*01 is also associated with rheumatoid arthritis and hematologic malignancies, all overrepresented in IBM and associating age-related stochastic accumulation of CD8+ CD28- T cells [1, 86]. HLA-DRB1 alleles expression also impacts durable control of viral replication, HLA DR B1*03:01 being associated with high HIV viremia [91, 92], while HLA DRB1*01 was associated with spontaneous viral clearance of hepatitis C [92].
HLA DRB1*13 is common for IBM susceptibility and for protection against infection with several viruses, including HIV, HCV, HBV [87]. In IBM, the HLA DRB1*13:01 was associated with the highest age of onset and the lower strength [88]. Nevertheless, intriguingly, HLA DRB1*13 was protective against autoimmune diseases such as systemic lupus erythematosus, psoriasis, systemic sclerosis, and others [93]. However, HLA DRB1*13 is also associated with a slow progression of HIV [94]. HLA DRB1 *13 is associated with the clearance of hepatitis B as well [95]. Surprisingly, HLA DRB1*13 is neuroprotective, along with apoE, against age-related brain changes [96]. HLA-C*14:02:01 allele was higher in IBM patients with high LGL T cell expression [84]. HLA-C*14:02 allele was also associated with a T cell response in HIV-1 infection, which was nevertheless non-protective for the viral infection [97].
HLA-F, found in IBM and Sjogren’s syndrome, also elicits antiviral responses through activation of the KIR3DS1+ NK cells [98, 99].
A bioinformatic analysis identified 10 genes in IBM, most of them involved in immune mediated and infectious diseases, including CCR5 (encoding the human C–C motif chemokine receptor type 5), IRF8 (interferon regulatory factor 8), HLA DRB1, CD74, and others [100]. CCR5 is also common for IBM susceptibility and for antiviral protection [88]. CCR5, expressed by tissue-resident memory T cells, is centrally involved in immunosurveillance, in inflammatory, autoimmune, and neoplastic disorders [101]. CCR5 also serves as an HIV co-receptor [102]. Similarly, a bioinformatic analysis found common molecular mechanisms between IBM and Sjogren’s syndrome, related to viral infection and antigen processing/presentation [99]. Amongst the 29 common genes identified, PSMB9 encodes the immunoproteasome B9, while CD74 encodes the cluster of differentiation 74 (also called HLA class II invariant gamma chain), a transmembrane glycoprotein contributing to antigen presentation [98]. CD74, as a key molecule of macrophage activation, involved in IFN-I and IFN-γ associated pathways in IBM and in the interaction between myofibers and macrophages in IBM [103]. CD74 interacts with the macrophage migration inhibitory factor, and CD74 upregulation contributes to immune damage during HIV infection [104].
To conclude, many genes predisposing to IBM are also involved in antiviral defense, mostly in generating interferon type I and type II.
Therapeutic strategies involving TDP-43 in IBM
IBM currently has no effective long-term therapy [1]. Immunosuppression later during the disease course did not improve IBM, and T cell depletion did not prevent vacuole formation and disease progression [10]. Moreover, immunosuppressive therapies may sometimes reveal an underlying chronic infection [20]. Therefore, HIV testing is advisable mostly in PM/IBM overlaps [20]. Also, pan-JAK inhibitors in aged mice alleviated the senescence—associated secretory phenotype but may also reactivate latent viruses [55]. Trials of immunosuppressive therapies in IBM have been recently nicely reviewed [105]. Immunosuppression is not routinely advised unless IBM is rapidly progressive or associated with other autoimmune diseases [105].
Followed both inflammatory and myodegenerative pathways presumed to be involved in IBM pathogenesis [105]. Most studies addressed inflammation or the involvement of T cells. Alemtuzumab (against CD52), natalizumab, anti-TNF alpha such as infliximab or etanercept, or IL-1 inhibitors as anakinra and canakinumab showed modest or no improvement [105], Rapamycin (sirolimus) targets mTOR important in IL-2 immune responses and protein metabolism (NCT04789070) [105]. Novel therapeutic avenues involve anti-KLRG1 antibodies, targeting a surface marker of the highly differentiated CD8T cells (NCT04659031) [84, 105]. Moreover, in HIV, the KLRG1 expression on NK cells correlates with HIV transcription, and targeting KLRG1 on NK cells potentially aids in elimination of HIV-infected cells [106]. Therapies against myodegeneration have recently become targets in clinical trials (arimoclomol, bimagrumab, follistatin, oxandrolone, rapamycin) [105].
Possible future directions may address other pathways. The attempts to reduce TDP-43 level led to muscle weakness and defective regeneration in myopathy models [6]. However, in neurological disorders such as ALS and other TDP-43-associated diseases, affecting skeletal and cardiac muscles besides neurons, there are several TDP-43 directed therapies [107, 108]. In ALS inhibition or deletion of cGAS and STING prevents TDP-43-induced upregulation of NF-kB and IFN type I [107]. Nevertheless, the neurological and muscular effects are not completely superposable [6].
Research including new therapies and repurposing for IBM some drugs used with other indications could serve as directions for the future [109]. Future therapeutic approaches could include inhibition of TDP-43 aggregation, the TDP-43-mitochondria association, proteasomal degradation of cytoplasmic TDP-43, or reducing TDP-43 aggregation-induced cell stress [37, 38, 110, 111]. Drugs stimulating the proteasome, such as chlorpromazine and other phenothiazines, methylene blue as a structural analogue of chlorpromazine and pyrazolones may target proteotoxic disorders [112]. The efficacy of zetomipzomib (KZR-616), a selective inhibitor of the immunoproteasome, is being studied in a phase 2 controlled multicenter study for active PM and DM [113, 114]. GSK3 inhibition decreases TDP-43 aggregation [34]. Lithium inhibits GSK-3 and induces autophagy, which may be relevant for IBM [115]. Also, lithium protected synapses from HIV-1 Tat-induced neuronal loss, in cultures and may be neuroprotective in HIV [116, 117]. Some other GSK3 inhibitors (including famotidine, naproxen, olanzapine, curcumin-all sterically hindering the enzyme binding pocket) may be tested for repurposing in IBM [33]. Also, regulating CYLD could be tested as a possible a therapeutic strategy in IBM [63].
The connection between a chronic viral infection and IBM deserves to be investigated further. There are questions waiting to be answered. Which factors are involved in transforming acute viral myositis into chronic inflammatory idiopathic myopathy? And moreover, why do some aged patients develop after a viral infection an IIM, for instance an anti-synthetase syndrome, and others an IBM? For instance, in HIV infection, what conditionate the switch from a PM phenotype to an IBM one? [4]. Serial studies in patients with chronic viral infections and signs of myopathy and/or sarcopenia would probably shed light on this progression, also regarding the progression to immunosenescence, mitochondrial dysfunction and proteinopathy, and the role of TDP-43 in this setting.
Conclusions
TDP-43 is important in preventing the dsRNA-induced IFN responses [13]. Viral infections may disrupt TDP-43 solubility and function, leading to its accumulation and lack of splicing regulation. The phenotypic differences between several IBM subtypes may be conditioned, besides genetic predisposing factors and age, also by environmental triggers such as certain viruses, and by epigenetic regulators [65]. Malat1 upregulation in certain viral infections may contribute to a protracted immune response [80].
Finding early disease markers and untangling mechanisms after a viral injury could inform whether there is a window of opportunity for the anti-inflammatory therapy, hopefully stopping or slowing the plethora of accompanying proteostasis, mitochondrial, and metabolic defects. Certain viruses, high viremia, coinfections, reactivation of latent viruses, and post-acute expansion of cytotoxic T cells may all contribute to IBM, mainly in an age-shaped immune landscape, with CD8+ T cells with IFN-ƴ production. In most such cases, the virally induced senescent, IFN-ƴ producing cytotoxic CD8+ T cells are the ones involved in IBM, in a genetically predisposed host. Immunophenotyping IBM patients to identify elevated CD8+ CD57+ populations may help stratify patients with prognostic and possibly therapeutic implications [84]. Identifying pathogenic mechanisms may lead to the identification of potential new treatments or to drug repurposing to improve the outcome in this debilitating disease.
Acknowledgements
Not applicable.
Abbreviations
- aa
Amino acid
- ALS
Amyotrophic lateral sclerosis
- BRCA1
Breast cancer susceptibility gene 1
- Anti-cN1A
Antibodies against the cytosolic 5′-nucleotidase 1A
- CANDLE syndrome
Chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature
- cGAS
Cyclic GMP–AMP synthase
- CHCHD10
Coiled-coil-helix-coiled-coil-helix domain-containing protein 10
- CMV
Cytomegalovirus
- CYLD
Cylindromatosis, a deubiquitinating enzyme that negatively regulates signal transduction pathways, such as NF-kB signaling pathways
- DM
Dermatomyositis
- dsRNA
Double-stranded RNA
- EBV
Epstein–Barr virus
- ERV
Endogenous retroviruses
- GBP2
Guanylate-binding protein 2
- GSK3
Glycogen synthase kinase 3
- HCV
Hepatitis C virus
- HIV
Human immunodeficiency virus
- hnRNP
Heterogeneous nuclear ribonucleoprotein
- HSPs
Heat shock proteins
- HTLV1
Human T-cell leukemia virus type 1
- IFN
Interferon
- IBM
Inclusion body myositis
- IIMs
Idiopathic inflammatory myopathies
- iPS
Immunoproteasomes
- IRF
Interferon regulatory factor
- lncRNA
Long non-coding RNA
- Malat1/MALAT1
Metastasis-associated lung adenocarcinoma transcript-1
- MDA-5
Melanoma differentiation-associated protein 5
- MHC
Major histocompatibility complex
- miRNA
MicroRNA
- mRNA
Messenger RNA
- NBR1
Neighbor of BRCA1
- NF-kB
Nuclear factor kappa B
- NK
Natural killer cells
- NLRP3
NOD-, LRR-, and pyrin domain-containing protein 3
- PASC
Post-acute sequelae SARS-CoV-2 infection
- PM
Polymyositis
- PSMB8
Proteasome subunit beta type-8
- Rbck1
RanBP-type and C3HC4-type zinc finger-containing protein 1
- RBP
RNA-binding proteins
- RIG-I
Retinoic acid-inducible gene-I
- RNA
Ribonucleic acid
- RRM
RNA-recognition motif
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SARS-CoV2 S1 RBD
SARS-CoV-2 spike S1 protein receptor binding domain
- SAMHD1
Sterile alpha motif domain and histidine-aspartate domain-containing protein 1
- SGs
Stress granules
- ssRNA
Single-stranded RNA
- STAT
Signal transducer and activator of transcription
- STING
Stimulator of interferon genes
- TARDBP
TAR-DNA-binding-protein 43 (transactive response DNA-binding protein of 43 kDa)
- TDP-43
TAR-DNA-binding protein 43
- TEMRA
Effector memory T cells re-expressing CD45RA
- TRIM21
Tripartite motif containing 21
- UPS
Ubiquitin-proteasome system
- YB
Y-box-binding protein-1
Author contributions
Conceptualization was presented by LD; documentation was collected by VV, AC, and RV; imaging was provided by RV and LD; writing—original draft preparation was revised by LD; writing—review and editing were prepared by RV, AC, and VV. All authors have read and agreed to the published version of the manuscript.
Funding
Partial support for open access publication has been received from Foundation Vital Team.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vitalie Văcăraş and Romana Vulturar have contributed equally to this work.
References
- 1.Greenberg SA. Inclusion body myositis: clinical features and pathogenesis. Nat Rev Rheumatol. 2019;15(5):257–72. 10.1038/s41584-019-0186-x. [DOI] [PubMed] [Google Scholar]
- 2.McLeish E, Slater N, Sooda A, Wilson A, Coudert JD, Lloyd TE, Needham M. Inclusion body myositis: the interplay between ageing, muscle degeneration and autoimmunity. Best Pract Res Clin Rheumatol. 2022;36(2):101761. 10.1016/j.berh.2022.101761. [DOI] [PubMed] [Google Scholar]
- 3.Lundberg IE, de Visser M, Werth VP. Classification of myositis. Nat Rev Rheumatol. 2018;14(5):269–78. 10.1038/nrrheum.2018.41. [DOI] [PubMed] [Google Scholar]
- 4.Nelke C, Kleefeld F, Preusse C, Ruck T, Stenzel W. Inclusion body myositis and associated diseases: an argument for shared immune pathologies. Acta Neuropathol Commun. 2022;10(1):84. 10.1186/s40478-022-01389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snedden AM, Kellett KAB, Lilleker JB, Hooper NM, Chinoy H. The role of protein aggregation in the pathogenesis of inclusion body myositis. Clin Exp Rheumatol. 2022;40(2):414–24. 10.55563/clinexprheumatol/pp0oso. [DOI] [PubMed] [Google Scholar]
- 6.Versluys L, Ervilha Pereira P, Schuermans N, De Paepe B, De Bleecker JL, Bogaert E, Dermaut B. Expanding the TDP-43 proteinopathy pathway from neurons to muscle: physiological and pathophysiological functions. Front Neurosci. 2022;16:815765. 10.3389/fnins.2022.815765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weihl CC, Temiz P, Miller SE, Watts G, Smith C, Forman M, Hanson PI, et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry. 2008;79(10):1186–9. 10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Šušnjar U, Škrabar N, Brown AL, Abbassi Y, Phatnani H, NYGC ALS Consortium, Cortese A, et al. Cell environment shapes TDP-43 function with implications in neuronal and muscle disease. Commun Biol. 2022;5(1):314. 10.1038/s42003-022-03253-8. [DOI] [PMC free article] [PubMed]
- 9.Buratti E. TDP-43 post-translational modifications in health and disease. Expert Opin Ther Targets. 2018;22(3):279–93. 10.1080/14728222.2018.1439923. [DOI] [PubMed] [Google Scholar]
- 10.Britson KA, Ling JP, Braunstein KE, Montagne JM, Kastenschmidt JM, Wilson A, Ikenaga C, et al. Loss of TDP-43 function and rimmed vacuoles persist after T cell depletion in a xenograft model of sporadic inclusion body myositis. Sci Transl Med. 2022;14(628):eabi9196. 10.1126/scitranslmed.abi9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve. 2009;40(1):19–31. 10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahic Z, Buratti E, Cappelli S. Reviewing the potential links between viral infections and TDP-43 proteinopathies. Int J Mol Sci. 2023;24(2):1581. 10.3390/ijms24021581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunker W, Ye X, Zhao Y, Liu L, Richardson A, Karijolich J. TDP-43 prevents endogenous RNAs from triggering a lethal RIG-I-dependent interferon response. Cell Rep. 2021;35(2):108976. 10.1016/j.celrep.2021.108976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Wang Z, Liu L, Yang Z, Liu S, Ma Z, Liu Y, et al. LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity. Proc Natl Acad Sci U S A. 2020;117(38):23695–706. 10.1073/pnas.2003932117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ylä-Anttila P. Autophagy receptors as viral targets. Cell Mol Biol Lett. 2021;26(1):29. 10.1186/s11658-021-00272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Askanas V, Engel WK. Inclusion-body myositis: newest concepts of pathogenesis and relation to aging and Alzheimer disease. J Neuropathol Exp Neurol. 2001;60(1):1–14. 10.1093/jnen/60.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Dubowsky M, Theunissen F, Carr JM, Rogers ML. The molecular link between TDP-43, endogenous retroviruses and inflammatory neurodegeneration in amyotrophic lateral sclerosis: a potential target for Triumeq, an antiretroviral therapy. Mol Neurobiol. 2023;60(11):6330–45. 10.1007/s12035-023-03472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morosetti R, Broccolini A, Sancricca C, Gliubizzi C, Gidaro T, Tonali PA, Ricci E, Mirabella M. Increased aging in primary muscle cultures of sporadic inclusion-body myositis. Neurobiol Aging. 2010;31(7):1205–14. 10.1016/j.neurobiolaging.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Germain MA, Chatel-Chaix L, Gagné B, Bonneil É, Thibault P, Pradezynski F, de Chassey B, et al. Elucidating novel hepatitis C virus-host interactions using combined mass spectrometry and functional genomics approaches. Mol Cell Proteomics. 2014;13(1):184–203. 10.1074/mcp.M113.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd TE, Pinal-Fernandez I, Michelle EH, Christopher-Stine L, Pak K, Sacktor N, Mammen AL. Overlapping features of polymyositis and inclusion body myositis in HIV-infected patients. Neurology. 2017;88(15):1454–60. 10.1212/WNL.0000000000003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saldi TK, Ash PE, Wilson G, Gonzales P, Garrido-Lecca A, Roberts CM, Dostal V, et al. TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. EMBO J. 2014;33(24):2947–66. 10.15252/embj.201488740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fung G, Shi J, Deng H, Hou J, Wang C, Hong A, Zhang J, et al. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ. 2015;22(12):2087–97. 10.1038/cdd.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rathore A, Iketani S, Wang P, Jia M, Sahi V, Ho DD. CRISPR-based gene knockout screens reveal deubiquitinases involved in HIV-1 latency in two Jurkat cell models. Sci Rep. 2020;10(1):5350. 10.1038/s41598-020-62375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Lu S, Gu J, Xia W, Zhang S, Zhang S, Wang Y, et al. SARS-CoV-2 impairs the disassembly of stress granules and promotes ALS-associated amyloid aggregation. Protein Cell. 2022;13(8):602–14. 10.1007/s13238-022-00905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Zhang J, Ke J, Zeng L, Cheng K, Han X, Chen F, et al. LncGBP9 knockdown alleviates myocardial inflammation and apoptosis in mice with acute viral myocarditis via suppressing NF-kappaB signaling pathamianway. Inflamm Res. 2022;71(12):1559–76. 10.1007/s00011-022-01644-5. [DOI] [PubMed] [Google Scholar]
- 26.Chang YH, Dubnau J. Endogenous retroviruses and TDP-43 proteinopathy form a sustaining feedback driving intercellular spread of Drosophila neurodegeneration. Nat Commun. 2023;14(1):966. 10.1038/s41467-023-36649-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller CW, Schmidt J, Lunemann JD. Immune and myodegenerative pathomechanisms in inclusion body myositis. Ann Clin Transl Neurol. 2017;4(6):422–45. 10.1002/acn3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishino H, Engel AG, Rima BK. Inclusion body myositis: the mumps virus hypothesis. Ann Neurol. 1989;25(3):260–4. 10.1002/ana.410250309. [DOI] [PubMed] [Google Scholar]
- 29.Agergaard J, Leth S, Pedersen TH, Harbo T, Blicher JU, Karlsson P, Ostergaard L, et al. Myopathic changes in patients with long-term fatigue after COVID-19. Clin Neurophysiol. 2021;132(8):1974–81. 10.1016/j.clinph.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galeotti C, Bayry J. Autoimmune and inflammatory diseases following COVID-19. Nat Rev Rheumatol. 2020;16(8):413–4. 10.1038/s41584-020-0448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen TM, Kabotyanski EB, Reineke LC, Shao J, Xiong F, Lee JH, Dubrulle J, et al. The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 2020;8(5):2621–42. 10.1093/nar/gkz1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li XL, Ezelle HJ, Hsi TY, Hassel BA. A central role for RNA in the induction and biological activities of type 1 interferons. Wiley Interdiscip Rev RNA. 2011;2(1):58–78. 10.1002/wrna.32. [DOI] [PubMed] [Google Scholar]
- 33.Piazzi M, Bavelloni A, Cenni V, Faenza I, Blalock WL. Revisiting the role of GSK3, a modulator of innate immunity, in idiopathic inclusion body myositis. Cells. 2021;10(11):3255. 10.3390/cells10113255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreedharan J, Neukomm LJ, Brown Jr RH, Freeman MR. Age-dependent TDP-43-mediated motor neuron degeneration requires GSK3, hat-trick, and xmas-2. Curr Biol. 2015;25(16):2130–6. 10.1016/j.cub.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu XL, Sun DD, Zheng MT, Li XT, Niu HH, Zhang L, Zhou ZW, et al. Maraviroc promotes recovery from traumatic brain injury in mice by suppression of neuroinflammation and activation of neurotoxic reactive astrocytes. Neural Regen Res. 2023;18(1):141–9. 10.4103/1673-5374.344829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medeiros GA, Silvério JC, Marino AP, Roffê E, Vieira V, Kroll-Palhares K, Carvalho CE, Silva AA, et al. Treatment of chronically Trypanosoma cruzi-infected mice with a CCR1/CCR5 antagonist (Met-RANTES) results in amelioration of cardiac tissue damage. Microbes Infect. 2009;11(2):264–73. 10.1016/j.micinf.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Webber CJ, Murphy CN, Rondón-Ortiz AN, van der Spek SJF, Kelly EX, Lampl NM, Chiesa G, et al. Human herpesvirus 8 ORF57 protein is able to reduce TDP-43 pathology: network analysis identifies interacting pathways. Hum Mol Genet. 2023;32(20):2966–80. 10.1093/hmg/ddad122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huntley ML, Gao J, Termsarasab P, Wang L, Zeng S, Thammongkolchai T, Liu Y, et al. Association between TDP-43 and mitochondria in inclusion body myositis. Lab Invest. 2019;99(7):1041–8. 10.1038/s41374-019-0233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Idrees D, Kumar V. SARS-CoV-2 spike protein interactions with amyloidogenic proteins: potential clues to neurodegeneration. Biochem Biophys Res Commun. 2021;554:94–8. 10.1016/j.bbrc.2021.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang K, Wang C, Vagts C, Raguveer V, Finn PW, Perkins DL. Long non-coding RNAs (lncRNAs) NEAT1 and MALAT1 are differentially expressed in severe COVID-19 patients: an integrated single-cell analysis. PLoS ONE. 2022;17(1):e0261242. 10.1371/journal.pone.0261242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menon MP, Hua KF. The long non-coding RNAs: paramount regulators of the NLRP3 inflammasome. Front Immunol. 2020;11:569524. 10.3389/fimmu.2020.569524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamann PD, Roux BT, Heward JA, Love S, McHugh NJ, Jones SW, Lindsay MA. Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis. Sci Rep. 2017;7(1):8024. 10.1038/s41598-017-08603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Askanas V, Engel WK, Nogalska A. Inclusion body myositis: a degenerative muscle disease associated with intra-muscle fiber multi-protein aggregates, proteasome inhibition, endoplasmic reticulum stress and decreased lysosomal degradation. Brain Pathol. 2009;19(3):493–506. 10.1111/j.1750-3639.2009.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghannam K, Martinez-Gamboa L, Spengler L, Krause S, Smiljanovic B, Bonin M, Bhattarai S, et al. Upregulation of immunoproteasome subunits in myositis indicates active inflammation with involvement of antigen presenting cells, CD8 T-cells and IFNGamma. PLoS ONE. 2014;9(8):e104048. 10.1371/journal.pone.0104048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Eshof BL, Medfai L, Nolfi E, Wawrzyniuk M, Sijts AJAM. The function of immunoproteasomes—an immunologists’ perspective. Cells. 2021;10(12):3360. 10.3390/cells10123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellington D, Yin Z, Yu Z, Heilig R, Davis S, Fischer R, Felce SL, et al. SARS-CoV-2 mutations affect antigen processing by the proteasome to alter CD8+ T cell responses. Heliyon. 2023;9(10):e20076. 10.1016/j.heliyon.2023.e20076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrer I, Martin B, Castano JG, Lucas JJ, Moreno D, Olive M. Proteasomal expression, induction of immunoproteasome subunits, and local MHC class I presentation in myofibrillar myopathy and inclusion body myositis. J Neuropathol Exp Neurol. 2004;63(5):484–98. 10.1093/jnen/63.5.484. [DOI] [PubMed] [Google Scholar]
- 48.Bolko L, Jiang W, Tawara N, Landon-Cardinal O, Anquetil C, Benveniste O, Allenbach Y. The role of interferons type I, II and III in myositis: a review. Brain Pathol. 2021;31(3):e12955. 10.1111/bpa.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanidze J, Hoffmann R, Lochmuller H, Engel AG, Hohlfeld R, Dornmair K. Inclusion body myositis: laser microdissection reveals differential up-regulation of IFN-gamma signaling cascade in attacked versus nonattacked myofibers. Am J Pathol. 2011;179(3):1347–59. 10.1016/j.ajpath.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ekholm L, Vosslamber S, Tjarnlund A, de Jong TD, Betteridge Z, McHugh N, Plestilova L, et al. Autoantibody specificities and type I interferon pathway activation in idiopathic inflammatory myopathies. Scand J Immunol. 2016;84(2):100–9. 10.1111/sji.12449. [DOI] [PubMed] [Google Scholar]
- 51.Angeles A, Fung G, Luo H. Immune and non-immune functions of the immunoproteasome. Front Biosci (Landmark Ed). 2012;17(5):1904–16. 10.1093/jnen/60.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Ayaki T, Murata K, Kanazawa N, Uruha A, Ohmura K, Sugie K, Kasagi S, et al. Myositis with sarcoplasmic inclusions in Nakajo-Nishimura syndrome: a genetic inflammatory myopathy. Neuropathol Appl Neurobiol. 2020;46(6):579–87. 10.1111/nan.12614. [DOI] [PubMed] [Google Scholar]
- 53.Pinal-Fernandez I, Casal-Dominguez M, Derfoul A, Pak K, Plotz P, Miller FW, Milisenda JC, et al. Identification of distinctive interferon gene signatures in different types of myositis. Neurology. 2019;93(12):e1193–204. 10.1212/WNL.0000000000008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rigolet M, Hou C, Baba Amer Y, Aouizerate J, Periou B, Gherardi RK, et al. Distinct interferon signatures stratify inflammatory and dysimmune myopathies. RMD Open. 2019;5(1):e000811. 10.1136/rmdopen-2018-000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goronzy JJ, Weyand CM. Mechanisms underlying T cell ageing. Nat Rev Immunol. 2019;19(9):573–83. 10.1038/s41577-019-0180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naddaf E, Shelly S, Mandrekar J, Chamberlain AM, Hoffman EM, Ernste FC, Liewluck T. Survival and associated comorbidities in inclusion body myositis. Rheumatology (Oxford). 2022;61(5):2016–24. 10.1093/rheumatology/keab716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goyal NA, Coulis G, Duarte J, Farahat PK, Mannaa AH, Cauchii J, Irani T, et al. Immunophenotyping of inclusion body myositis blood T and NK cells. Neurology. 2022;98(13):e1374–83. 10.1212/WNL.0000000000200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eloranta ML, Barbasso Helmers S, Ulfgren AK, Ronnblom L, Alm GV, Lundberg IE. A possible mechanism for endogenous activation of the type I interferon system in myositis patients with anti-Jo-1 or anti-Ro 52/anti-Ro 60 autoantibodies. Arthritis Rheum. 2007;56(9):3112–24. 10.1002/art.22860. [DOI] [PubMed] [Google Scholar]
- 59.Jones EL, Laidlaw SM, Dustin LB. TRIM21/Ro52—roles in innate immunity and autoimmune disease. Front Immunol. 2021;12:738473. 10.3389/fimmu.2021.738473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinal-Fernandez I, Casal-Dominguez M, Derfoul A, Pak K, Miller FW, Milisenda JC, Grau-Junyent JM. Machine learning algorithms reveal unique gene expression profiles in muscle biopsies from patients with different types of myositis. Ann Rheum Dis. 2020;79(9):1234–42. 10.1136/annrheumdis-2019-216599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai Y, Zhu Y, Zheng J, Zhang Y, Chen W. NBR1 mediates autophagic degradation of IRF3 to negatively regulate type I interferon production. Biochem Biophys Res Commun. 2022;623:140–7. 10.1016/j.bbrc.2022.07.043. [DOI] [PubMed] [Google Scholar]
- 62.D’Agostino C, Nogalska A, Cacciottolo M, Engel WK, Askanas V. Abnormalities of NBR1, a novel autophagy-associated protein, in muscle fibers of sporadic inclusion-body myositis. Acta Neuropathol. 2011;122(5):627–36. 10.1007/s00401-011-0874-3. [DOI] [PubMed] [Google Scholar]
- 63.Yamashita S, Matsuo Y, Tawara N, Hara K, Yamamoto M, Nishikami T, Kawakami K, et al. CYLD dysregulation in pathogenesis of sporadic inclusion body myositis. Sci Rep. 2019;9(1):11606. 10.1038/s41598-019-48115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Jia Q, Gao W, Zhang W. The role of deubiquitinases in virus replication and host innate immune response. Front Microbiol. 2022;13:839624. 10.3389/fmicb.2022.839624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L, Shorter J, et al. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 2019;102(2):339–57. 10.1016/j.neuron.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adler BL, Christopher-Stine L. Triggers of inflammatory myopathy: insights into pathogenesis. Discov Med. 2018;25(136):75–83. [PMC free article] [PubMed] [Google Scholar]
- 67.Singh H, Talapatra P, Arya S, Gupta V. Viral myositis as a close mimicker of polymyositis. Ann Trop Med Public Health. 2013;6:324–6. 10.4103/1755-6783.120997. [Google Scholar]
- 68.Manzano GS, Woods JK, Amato AA. Covid-19-associated myopathy caused by type I interferonopathy. N Engl J Med. 2020;383(24):2389–90. 10.1056/NEJMc2031085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uruha A, Noguchi S, Hayashi YK, Tsuburaya RS, Yonekawa T, Nonaka I, Nishino I. Hepatitis C virus infection in inclusion body myositis: a case-control study. Neurology. 2016;86(3):211–7. 10.1212/WNL.0000000000002291. [DOI] [PubMed] [Google Scholar]
- 70.Eura N. Anti-cytosolic 5’-nucleotidase 1A (cN1A) positivity in muscle is helpful in the diagnosis of sporadic inclusion body myositis: a study of 35 Japanese patients. J Neurol Neurosci. 2016. 10.3390/biomedicines11071963. [Google Scholar]
- 71.Matsuura E, Umehara F, Nose H, Higuchi I, Matsuoka E, Izumi K, Kubota R, et al. Inclusion body myositis associated with human T-lymphotropic virus-type I infection: eleven patients from an endemic area in Japan. J Neuropathol Exp Neurol. 2008;67(1):41–9. 10.1097/nen.0b013e31815f38b7. [DOI] [PubMed] [Google Scholar]
- 72.Ou SH, Wu F, Harrich D, García-Martínez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69(6):3584–96. 10.1128/JVI.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nehls J, Koppensteiner H, Brack-Werner R, Floss T, Schindler M. HIV-1 replication in human immune cells is independent of TAR DNA binding protein 43 (TDP-43) expression. PLoS ONE. 2014;9(8):e105478. 10.1371/journal.pone.0105478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou L, Miranda-Saksena M, Saksena NK. Viruses and neurodegeneration. Virol J. 2013;10:172. 10.1186/1743-422X-10-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dalal F, Dalal H, McNew G. COVID-19-induced sporadic inclusion body myositis. Cureus. 2022;14(10):e30808. 10.7759/cureus.30808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saud A, Naveen R, Aggarwal R, Gupta L. COVID-19 and myositis: what we know so far. Curr Rheumatol Rep. 2021;23(8):63. 10.1007/s11926-021-01023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jacob S, Kapadia R, Soule T, Luo H, Schellenberg KL, Douville RN, Pfeffer G. Neuromuscular complications of SARS-CoV-2 and other viral infections. Front Neurol. 2022;13:914411. 10.3389/fneur.2022.914411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swartzman I, Gu JJ, Toner Z, Grover R, Suresh L, Ullman LE. Prevalence of myositis-specific autoantibodies and myositis-associated autoantibodies in COVID-19 patients: a pilot study and literature review. Cureus. 2022;14(9):e29752. 10.7759/cureus.29752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holzer MT, Krusche M, Ruffer N, Haberstock H, Stephan M, Huber TB, Kotter I. New-onset dermatomyositis following SARS-CoV-2 infection and vaccination: a case-based review. Rheumatol Int. 2022;42(12):2267–76. 10.1007/s00296-022-05176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, Kang L, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397(10270):220–32. 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, Li S, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185(5):881–95. 10.1016/j.cell.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramakrishnan RK, Kashour T, Hamid Q, Halwani R, Tleyjeh IM. Unraveling the mystery surrounding post-acute sequelae of COVID-19. Front Immunol. 2021;12:686029. 10.3389/fimmu.2021.686029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang R, Roy B, Wu Q, Mohanty S, Nowak RJ, Shaw AC, Kleinstein SH, O’Connor KC. The plasma cell infiltrate populating the muscle tissue of patients with inclusion body myositis features distinct B cell receptor repertoire properties. Immunohorizons. 2023;7(5):310–22. 10.4049/immunohorizons.2200078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McLeish E, Sooda A, Slater N, Kachigunda B, Beer K, Paramalingam S, Lamont PJ, Chopra A, Mastaglia FL, Needham M, Coudert JD. Uncovering the significance of expanded CD8+ large granular lymphocytes in inclusion body myositis: Insights into T cell phenotype and functional alterations, and disease severity. Front Immunol. 2023;14:1153789. 10.3389/fimmu.2023.1153789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rose A, Isenalumhe L, Van den Bergh M, Sokol L. Clonal T-cell large granular lymphocytic disorders manifesting in patients with HIV-1 infection: case series and review of the literature. Mediterr J Hematol Infect Dis. 2018;10(1):e2018036. 10.4084/MJHID.2018.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goronzy JJ, Li G, Yang Z, Weyand CM. The Janus head of T cell aging—autoimmunity and immunodeficiency. Front Immunol. 2013;4:131. 10.3389/fimmu.2013.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rothwell S, Cooper RG, Lundberg IE, Gregersen PK, Hanna MG, Machado PM, Herbert MK, et al. Immune-array analysis in sporadic inclusion body myositis reveals HLA-DRB1 amino acid heterogeneity across the myositis spectrum. Arthritis Rheumatol. 2017;69(5):1090–9. 10.1002/art.40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rojana-Udomsart A, Bundell C, James I, Castley A, Martinez P, Christiansen F, Hollingsworth P, Mastaglia F. Frequency of autoantibodies and correlation with HLA-DRB1 genotype in sporadic inclusion body myositis (s-IBM): a population control study. J Neuroimmunol. 2012;249(1–2):66–70. 10.1016/j.jneuroim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 89.Slater N, Sooda A, McLeish E, Beer K, Brusch A, Shakya R, Bundell C, James I, Chopra A, Mastaglia FL, Needham M, Coudert JD. High-resolution HLA genotyping in inclusion body myositis refines 8.1 ancestral haplotype association to DRB1*03:01:01 and highlights pathogenic role of arginine-74 of DRβ1 chain. J Autoimmun. 2024;142:103150. 10.1016/j.jaut.2023.103150. [DOI] [PubMed] [Google Scholar]
- 90.Miller FW, Lamb JA, Schmidt J, Nagaraju K. Risk factors and disease mechanisms in myositis. Nat Rev Rheumatol. 2018;14(5):255–68. 10.1038/nrrheum.2018.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ranasinghe S, Cutler S, Davis I, Lu R, Soghoian DZ, Qi Y, Sidney J, Kranias G, Flanders MD, Lindqvist M, Kuhl B, Alter G, Deeks SG, Walker BD, Gao X, Sette A, Carrington M, Streeck H. Association of HLA-DRB1-restricted CD4+ T cell responses with HIV immune control. Nat Med. 2013;19(7):930–3. 10.1038/nm.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barrett S, Ryan E, Crowe J. Association of the HLA-DRB1*01 allele with spontaneous viral clearance in an Irish cohort infected with hepatitis C virus via contaminated anti-D immunoglobulin. J Hepatol. 1999;30(6):979–83. 10.1016/s0168-8278(99)80249-9. [DOI] [PubMed] [Google Scholar]
- 93.Bettencourt A, Carvalho C, Leal B, Brás S, Lopes D, Martins da Silva A, Santos E, et al. The protective role of HLA-DRB1(∗)13 in autoimmune diseases. J Immunol Res. 2015. 10.1155/2015/948723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferre AL, Hunt PW, McConnell DH, Morris MM, Garcia JC, Pollard RB, Yee HF, et al. HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses. J Virol. 2010;84(21):11020–9. 10.1128/JVI.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14(12):841–8. 10.1111/j.1365-2893.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- 96.James LM, Christova P, Lewis SM, Engdahl BE, Georgopoulos A, Georgopoulos AP. Protective effect of human leukocyte antigen (HLA) allele DRB1*13:02 on age-related brain gray matter volume reduction in healthy women. EBioMedicine. 2018;29:31–7. 10.1016/j.ebiom.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chikata T, Paes W, Kuse N, Partridge T, Gatanaga H, Zhang Y, Kuroki K, Maenaka K, Ternette N, Oka S, Borrow P, Takiguchi M. Impact of micropolymorphism outside the peptide binding groove in the clinically relevant allele HLA-C*14 on T cell responses in HIV-1 infection. J Virol. 2022;96(10):e0043222. 10.1128/jvi.00432-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zeng L, Chen K, Xiao F, Zhu CY, Bai JY, Tan S, Long L, et al. Potential common molecular mechanisms between Sjögren syndrome and inclusion body myositis: a bioinformatic analysis and in vivo validation. Front Immunol. 2023;14:1161476. 10.3389/fimmu.2023.1161476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin A, Yan WH. The emerging roles of human leukocyte antigen-F in immune modulation and viral infection. Front Immunol. 2019;10:964. 10.3389/fimmu.2019.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Khasanova E, Zhang L. Bioinformatics analysis of gene expression profiles of Inclusion body myositis. Scand J Immunol. 2020;91(6):e12887. 10.1111/sji.12887. [DOI] [PubMed] [Google Scholar]
- 101.Jasinska AJ, Pandrea I, Apetrei C. CCR5 as a coreceptor for human immunodeficiency virus and simian immunodeficiency viruses: a prototypic love-hate affair. Front Immunol. 2022;13:835994. 10.3389/fimmu.2022.835994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ellwanger JH, Kulmann-Leal B, Kaminski VL, Rodrigues AG, Bragatte MAS, Chies JAB. Beyond HIV infection: neglected and varied impacts of CCR5 and CCR5Δ32 on viral diseases. Virus Res. 2020;286:198040. 10.1016/j.virusres.2020.198040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roos A, Preusse C, Hathazi D, Goebel HH, Stenzel W. Proteomic profiling unravels a key role of specific macrophage subtypes in sporadic inclusion body myositis. Front Immunol. 2019;10:1040. 10.3389/fimmu.2019.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Trifone C, Salido J, Ruiz MJ, Leng L, Quiroga MF, Salomón H, Bucala R, et al. Interaction between macrophage migration inhibitory factor and CD74 in human immunodeficiency virus type I infected primary monocyte-derived macrophages triggers the production of proinflammatory mediators and enhances infection of unactivated CD4+ T cells. Front Immunol. 2018;9:1494. 10.3389/fimmu.2018.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Connolly CM, Plomp L, Paik JJ, Allenbach Y. Possible future avenues for myositis therapeutics: DM, IMNM and IBM. Best Pract Res Clin Rheumatol. 2022;36(2):101762. 10.1016/j.berh.2022.101762. [DOI] [PubMed] [Google Scholar]
- 106.Astorga-Gamaza A, Perea D, Sanchez-Gaona N, Calvet-Mirabent M, Gallego-Cortés A, Grau-Expósito J, Sanchez-Cerrillo I, Rey J, Castellví J, Curran A, Burgos J, Navarro J, Suanzes P, Falcó V, Genescà M, Martín-Gayo E, Buzon MJ. KLRG1 expression on natural killer cells is associated with HIV persistence, and its targeting promotes the reduction of the viral reservoir. Cell Rep Med. 2023;4(10):101202. 10.1016/j.xcrm.2023.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu CH, Davidson S, Harapas CR, Hilton JB, Mlodzianoski MJ, Laohamonthonkul P, Louis C, et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183(3):636–49. 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mori F, Tada M, Kon T, Miki Y, Tanji K, Kurotaki H, Tomiyama M, et al. Phosphorylated TDP-43 aggregates in skeletal and cardiac muscle are a marker of myogenic degeneration in amyotrophic lateral sclerosis and various conditions. Acta Neuropathol Commun. 2019;7(1):165. 10.1186/s40478-019-0824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Damian L, Login CC, Solomon C, Belizna C, Encica S, Urian L, Jurcut C, et al. Inclusion body myositis and neoplasia: a narrative review. Int J Mol Sci. 2022;23(13):7358. 10.3390/ijms23137358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pozzi S, Codron P, Soucy G, Renaud L, Cordeau PJ, Dutta K, Bareil C, Julien JP. Monoclonal full-length antibody against TAR DNA binding protein 43 reduces related proteinopathy in neurons. JCI Insight. 2020;5(21):e140420. 10.1172/jci.insight.140420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang P, Deng J, Dong J, Liu J, Bigio EH, Mesulam M, Wang T, et al. TDP-43 induces mitochondrial damage and activates the mitochondrial unfolded protein response. PLoS Genet. 2019;15(5):e1007947. 10.1371/journal.pgen.1007947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Njomen E, Tepe JJ. Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J Med Chem. 2019;62(14):6469–81. 10.1021/acs.jmedchem.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moghadam-Kia S, Oddis CV. Current and new targets for treating myositis. Curr Opin Pharmacol. 2022;65:102257. 10.1016/j.coph.2022.102257. [DOI] [PubMed] [Google Scholar]
- 114.Guglielmi V, Cheli M, Tonin P, Vattemi G. Sporadic inclusion body myositis at the crossroads between muscle degeneration, inflammation, and aging. Int J Mol Sci. 2024;25(5):2742. 10.3390/ijms25052742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Terracciano C, Nogalska A, Engel WK, Askanas V. In AbetaPP-overexpressing cultured human muscle fibers proteasome inhibition enhances phosphorylation of AbetaPP751 and GSK3beta activation: effects mitigated by lithium and apparently relevant to sporadic inclusion-body myositis. J Neurochem. 2010;112(2):389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hong N, Park JS, Kim HJ. Synapto-protective effect of lithium on HIV-1 Tat-induced synapse loss in rat hippocampal cultures. Anim Cells Syst (Seoul). 2021;26(1):1–9. 10.1080/19768354.2021.2018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schifitto G, Zhong J, Gill D, Peterson DR, Gaugh MD, Zhu T, Tivarus M, Cruttenden K, Maggirwar SB, Gendelman HE, Dewhurst S, Gelbard HA. Lithium therapy for human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol. 2009;15(2):176–86. 10.1080/13550280902758973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.