Analyses of longitudinal peripheral biomarkers and baseline tumor tissue from JAVELIN Renal 101 provide novel evidence of differential immunomodulatory mechanisms in patients with advanced renal cell carcinoma treated with avelumab plus axitinib versus sunitinib.
Abstract
The phase III JAVELIN Renal 101 trial demonstrated prolonged progression-free survival (PFS) in patients (N = 886) with advanced renal cell carcinoma treated with first-line avelumab + axitinib (A+Ax) versus sunitinib. We report novel findings from integrated analyses of longitudinal blood samples and baseline tumor tissue. PFS was associated with elevated lymphocyte levels in the sunitinib arm and an abundance of innate immune subsets in the A+Ax arm. Treatment with A+Ax led to greater T-cell repertoire modulation and less change in T-cell numbers versus sunitinib. In the A+Ax arm, patients with tumors harboring mutations in ≥2 of 10 previously identified PFS-associated genes (double mutants) had distinct circulating and tumor-infiltrating immunologic profiles versus those with wild-type or single-mutant tumors, suggesting a role for non–T-cell–mediated and non–natural killer cell–mediated mechanisms in double-mutant tumors. We provide evidence for different immunomodulatory mechanisms based on treatment (A+Ax vs. sunitinib) and tumor molecular subtypes.
Significance:
Our findings provide novel insights into the different immunomodulatory mechanisms governing responses in patients treated with avelumab (PD-L1 inhibitor) + axitinib or sunitinib (both VEGF inhibitors), highlighting the contribution of tumor biology to the complexity of the roles and interactions of infiltrating immune cells in response to these treatment regimens.
This article is featured in Selected Articles from This Issue, p. 384
INTRODUCTION
In the phase III JAVELIN Renal 101 trial, first-line (1L) treatment with avelumab, an anti–programmed cell death 1 ligand 1 (PD-L1) immune-checkpoint inhibitor (ICI), plus axitinib (A+Ax), a tyrosine kinase inhibitor that targets vascular endothelial growth factor (VEGF) receptors 1, 2, and 3, significantly improved progression-free survival (PFS) and objective response rate compared with sunitinib, a multitargeted VEGF tyrosine kinase inhibitor, in patients with advanced renal cell carcinoma (aRCC; refs. 1–4). Follow-up for overall survival is ongoing. Consequently, treatment with 1L A+Ax has received regulatory approval in several countries worldwide (3–6).
A previous report explored the biological mechanisms underlying the clinical activity of A+Ax and summarized results from a range of molecular analyses using pretreatment tumor samples from JAVELIN Renal 101 (7). Novel immune and angiogenesis gene-expression signatures (GES), called “JAVELIN Renal 101 signatures,” were identified to be associated with efficacy outcomes in the A+Ax and sunitinib treatment arms (7). The immune GES included 6 genes involved in natural killer (NK) cell–mediated cytotoxicity (CD2, CD96, PRF1, CD244, KLRD1, and SH2D1A; ref. 7). This was consistent with in vitro studies showing that, in addition to the conventional mechanism of cytotoxic T-cell activity associated with PD-(L)1 inhibitors, the intact Fc region of the IgG1 avelumab antibody also enables antibody-dependent cell-mediated cytotoxicity (ADCC) against tumor cells expressing PD-L1 on their surface (8). In addition, recent molecular analyses from the JAVELIN Bladder 100 trial of avelumab 1L maintenance in patients with advanced urothelial cancer showed longer overall survival in a subgroup of patients with ≥2 FcγR alleles with high affinity for the IgG1 isotype, as well as higher levels of innate and adaptive immune cells in tumors (9). However, an association with the high-affinity FcγR alleles was not observed in JAVELIN Renal 101, though a recent study from the JAVELIN Renal 101 trial examined sarcomatoid features in patient tumors and identified attributes that differentiate sarcomatoid RCC (sRCC) from non-sarcomatoid lesions, as well as features of sRCC that are associated with improved outcomes with A+Ax treatment (7, 10). We previously used a series of Cox proportional hazards models to examine the somatic variants from JAVELIN Renal 101 pretreatment tumor tissue and identified a set of 10 genes that, when mutated, were independently associated with prolonged PFS in the A+Ax arm (CD163L1, DNMT1, IL16, SPATA31C2, MYH7B, STAB2, CROCC2/LOC728763, FOXO1, ABCA1, and MC1R; ref. 7). Building on these data, we found that a double-mutant (DM) tumor genotype (variations in ≥2 of the 10 identified genes) was associated with longer PFS in patients treated with A+Ax but shorter PFS in those treated with sunitinib, suggesting a unique phenotype in patients with DM tumors (7).
Published analyses of peripheral analytes in aRCC are limited. In the JAVELIN Renal 101 trial, samples were collected from the entire trial population (N = 886), representing one of the largest data sets for aRCC to date and enabling pretreatment and on-treatment analyses of peripheral blood-based biomarkers, such as circulating cell subsets, T-cell receptors (TCR), and soluble analytes. Here, we report findings from integrated exploratory analyses of these data sets, including circulating proteins, cell populations, and the TCR repertoire, and assess their association with patient outcomes in the context of pretreatment tumor characteristics, including the presence of tumor-infiltrating leukocytes (TIL) and tumor genotype. Overall, our findings demonstrated different immunomodulatory mechanisms in patients treated with A+Ax versus sunitinib and in subsets of patients with different tumor microenvironments and genotypes.
RESULTS
Profiling of Circulating Cell Populations, Cytokines, and Chemokines
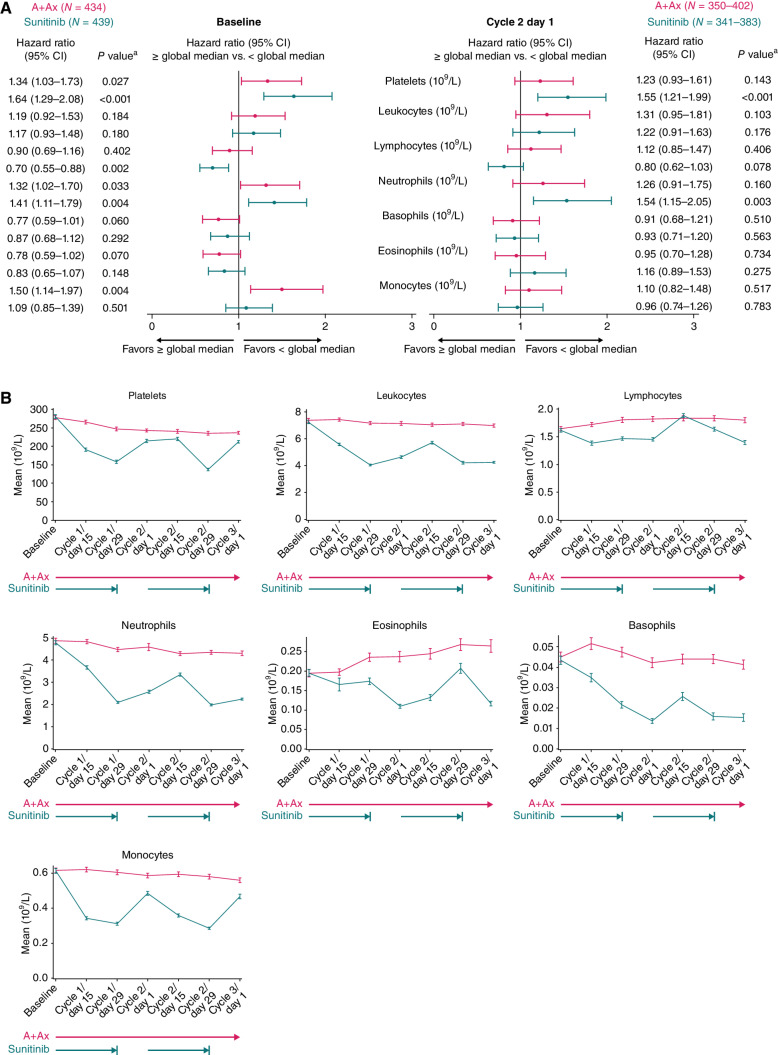
Peripheral cell populations and common blood-based biomarkers were analyzed in baseline and on-treatment samples (Fig. 1; Supplementary Figs. S1 and S2). In analyses of both treatment arms combined, selected peripheral cell populations showed potential prognostic effects, consistent with those seen in previous studies in aRCC (11, 12); longer PFS was associated with low (< median) baseline levels of circulating platelets, neutrophils, and monocytes [HR, 1.48 (95% CI, 1.24–1.76); P < 0.001; HR, 1.38 (95% CI, 1.16–1.65); P < 0.001; and HR, 1.25 (95% CI, 1.05–1.51); P = 0.015, respectively] and high (≥ median) baseline levels of lymphocytes and eosinophils [HR, 0.80 (95% CI, 0.67–0.95); P = 0.012 and HR, 0.81 (95% CI, 0.68–0.98); P = 0.027, respectively; Supplementary Fig. S1]. When circulating cells were examined in the A+Ax and sunitinib arms separately, longer PFS was associated with lower baseline levels of platelets and neutrophils, with numerically larger HRs observed in the sunitinib arm [A+Ax: HR, 1.34 (1.03–1.73); P = 0.027 and HR, 1.32 (1.02–1.70); P = 0.033, respectively; sunitinib: HR, 1.64 (1.29–2.08); P < 0.001 and HR, 1.41 (1.11–1.79); P = 0.004, respectively; Fig. 1A]. In the A+Ax arm, longer PFS was also associated with low baseline levels of monocytes [HR, 1.50 (95% CI, 1.14–1.97); P = 0.004], which was not seen with sunitinib [HR, 1.09 (95% CI, 0.85–1.39); P = 0.501]. In the sunitinib arm, longer PFS was associated with high levels of lymphocytes [HR, 0.70 (95% CI, 0.55–0.88); P = 0.002], which was not seen with A+Ax [HR, 0.90 (95% CI, 0.69–1.16); P = 0.402]. Associations of the neutrophil-to-lymphocyte ratio with PFS in the JAVELIN Renal 101 trial have been previously described (13).
Figure 1.
Association between PFS and circulating cell population numbers (109 cells/L) at baseline and following treatment with A+Ax or sunitinib. A, PFS according to numbers of different cell populations at baseline (left) and cycle 2 day 1 (right). For cycle 2 day 1, N values vary by subset; minimum to maximum N values are shown. Hazard ratios were calculated using the Cox proportional hazards model with < median used as the reference group. An HR of <1 indicates longer PFS in the ≥ median subgroup; an HR of >1 indicates longer PFS in the < median subgroup. B, Mean numbers of different peripheral cell populations at different time points. Error bars show standard error of the mean. Pink and teal arrows represent treatment with A+Ax and sunitinib, respectively. aTwo-sided P value comparing median cutoff subgroups (log-rank test); a statistical threshold value of P < 0.05 highlights observations of likely biological relevance.
Levels of these peripheral cell populations during treatment (through cycle 3 day 1) exhibited little change relative to baseline in the A+Ax arm. In contrast, all populations in the sunitinib arm demonstrated greater fluctuations relative to baseline, and these oscillations mirrored the sunitinib dosing schedule (4 weeks on treatment, 2 weeks off treatment; Fig. 1B). In the A+Ax arm, PFS was not associated with on-treatment levels (cycle 2 day 1) of peripheral blood cell types as was observed for baseline (i.e., platelets, neutrophils, and monocytes; Fig. 1A). In the sunitinib arm, longer PFS was associated with low levels of neutrophils and platelets during treatment [HR, 1.54 (95% CI, 1.15–2.05); P = 0.003 and HR, 1.55 (95% CI, 1.21–1.99); P < 0.001, respectively]; however, the baseline association between PFS and lymphocyte numbers was weaker during treatment. Despite this weaker association, the lymphocyte subset was the only cell type with a noteworthy treatment-by-biomarker interaction during treatment in patients in the sunitinib arm (Cox proportional hazards 2-sided P = 0.073).
Previous studies in aRCC and other malignancies have described that poor efficacy outcomes are associated with high levels of systemic inflammatory markers (14–17). Similarly, in both our treatment arms, a shorter PFS was associated with high baseline levels of systemic inflammatory markers, including interleukin (IL)-6, IL8, IL10, and C-reactive protein (CRP), with larger HRs observed for IL6 and IL10 in the sunitinib arm (Cox proportional hazards 2-sided P = 0.036 and P = 0.017 for IL6 and IL10 interactions, respectively); a similar pattern was observed at cycle 2 day 1 (Supplementary Fig. S2).
T-cell Receptor Repertoire Analyses
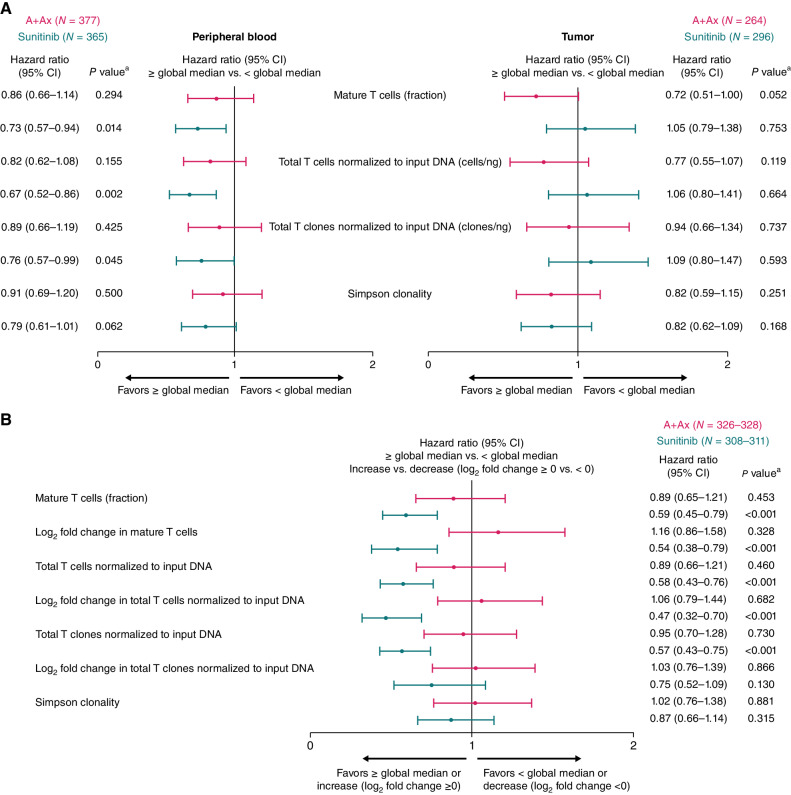
T-cell numbers and repertoires were characterized via core quantitation and repertoire metrics using bulk TCR repertoire sequencing (TCR-seq) in pretreatment tumor specimens and peripheral blood at baseline (Fig. 2A) and in peripheral blood during treatment (Fig. 2B). In the sunitinib arm, baseline peripheral T-cell quantitation measures describing a higher fraction and normalized number of T cells and clones in peripheral blood were associated with longer PFS [HR, 0.73 (95% CI, 0.57–0.94); P = 0.014; HR, 0.67 (95% CI, 0.52–0.86); P = 0.002; and HR, 0.76 (95% CI, 0.57–0.99); P = 0.045, respectively; Fig 2A]. However, although analyses in the A+Ax arm showed similar trends, no associations with PFS were observed in peripheral blood, though a strong trend toward longer PFS was observed with a higher fraction of infiltrating T cells in the pretreatment tumor in the A+Ax arm but not in the sunitinib arm (Fig. 2A). In the sunitinib arm, the peripheral T-cell population had expanded by the end of cycle 1, with median log2 fold change in T-cell quantitation measures from baseline to cycle 2 day 1 ranging from 0.364 to 0.449 and resulting in markedly higher levels versus the A+Ax arm (P < 0.0001; Table 1). Furthermore, in the sunitinib arm, high peripheral T-cell quantitation measures during treatment and an increase versus baseline (log2 fold change ≥0) were both associated with longer PFS; however, these metrics were not associated with PFS in the A+Ax arm (Fig. 2B). Consequently, an interaction of these measures with sunitinib treatment was observed for most T-cell metrics. These sunitinib treatment–by-biomarker interactions included the peripheral T-cell fraction, normalized T cells, and normalized T-cell clones (≥ vs. < global median; T-cell fraction 2-sided P = 0.059; normalized T cells P = 0.038; normalized clones P = 0.013) and increase versus decrease in T-cell fraction and normalized T cells (log2 fold change ≥0 vs. <0; T-cell fraction 2-sided P = 0.002; normalized T cells P = 0.001).
Figure 2.
PFS according to T-cell quantitation or repertoire metrics. A, PFS according to T-cell repertoire metrics in baseline peripheral blood (left) or pretreatment tumor samples (right). B, PFS according to T-cell repertoire metrics in peripheral blood during treatment (cycle 2 day 1) with A+Ax or sunitinib. Hazard ratios were calculated using the Cox proportional hazards model with < median or decrease (log2 fold change <0) used as the reference group. An HR of <1 indicates longer PFS in the ≥ median or increase (log2 fold change ≥0) subgroup, whereas an HR of >1 indicates longer PFS in the < median or decrease (log2 fold change <0) subgroup. aTwo-sided P value comparing median or cutoff log2 fold change subgroups (log-rank test); a statistical threshold value of P < 0.05 highlights observations of likely biological relevance.
Table 1.
Parameters related to T-cell receptor quantitation and repertoire change metrics during treatment.
| A+Ax (n = 328)a median (95% CI) |
Sunitinib (n = 311)a median (95% CI) |
P valueb | |
|---|---|---|---|
| Peripheral T-cell quantitation at cycle 2 day 1 | |||
| Mature T-cell fraction | 0.199 (0.188–0.211) | 0.261 (0.250–0.271) | <0.0001 |
| Log2 fold change in mature T-cell fraction | 0.092 (0.044–0.133) | 0.449 (0.369–0.504) | <0.0001 |
| Total T-cell clones (normalized to input DNA) | 0.792 (0.708–0.851) | 1.035 (0.955–1.073) | <0.0001 |
| Log2 fold change in total T-cell clones (normalized to input DNA) | 0.046 (0.022–0.073) | 0.364 (0.300–0.440) | <0.0001 |
| Total T cells (normalized to input DNA) | 27.310 (25.832–28.790) | 36.069 (34.860–38.031) | <0.0001 |
| Log2 fold change in total T cells (normalized to input DNA) | 0.079 (0.025–0.108) | 0.421 (0.358–0.486) | <0.0001 |
| Change in peripheral T-cell repertoire at cycle 2 day 1 vs. baseline | |||
| Log2 fold change in Simpson clonality | 0.117 (0.076–0.149) | 0.034 (0.000–0.069) | 0.0028 |
| Morisita index | 0.950 (0.943–0.960) | 0.966 (0.960–0.972) | 0.0015 |
| No. of expanded peripheral clones (beta-binomial) | 9.0 (7.0–10.0) | 5.0 (4.0–6.0) | <0.0001 |
| No. of contracted peripheral clones (beta-binomial) | 3.0 (2.0–4.0) | 2.0 (2.0–3.0) | 0.0018 |
| Maximum clone expansion | 0.006 (0.005–0.007) | 0.004 (0.003–0.005) | 0.0005 |
| Maximum clone contraction | −0.002 (−0.003, −0.002) | −0.003 (−0.003, −0.002) | 0.6218 |
aA+Ax n varies from 326 to 328 by metric; sunitinib n varies from 308 to 311 by metric.
bTwo-sided P value is based on the Wilcoxon rank sum test; a statistical threshold value of P < 0.05 highlights observations of likely biological relevance.
Although changes in T-cell quantitation measures from baseline to cycle 2 day 1 with A+Ax were relatively small (median log2 fold changes ranging from 0.046–0.092), metrics describing the change in repertoire composition between time points showed greater modulation with A+Ax than with sunitinib, including a lower Morisita index, higher maximum clone expansion value, a greater number of expanded and contracted clones using the beta-binomial model, and greater log2 fold change in Simpson clonality (Table 1). Changes in the repertoire were not associated with PFS in either arm; however, in the sunitinib arm, a numerically longer PFS was observed in patients with less repertoire modulation [≥ median Morisita index; HR vs. < median Morisita index, 0.78 (95% CI, 0.59–1.02); P = 0.070; Supplementary Fig. S3].
Analyses of Mutation-Defined Tumor Molecular Subgroups: DM Tumors versus Others
Previous translational analyses of pretreatment tumor samples from JAVELIN Renal 101 identified 10 genes for which somatic mutations or germline variants were associated with differential PFS (7). In the A+Ax arm, longer PFS was observed in patients whose tumors harbored mutations in ≥2 of these 10 genes (DM tumors) versus those with tumors harboring 0 or 1 mutation [termed wild-type (WT) and single (WT/S) mutants]; however, this was not observed in the sunitinib arm. These 10 genes are functionally diverse, and it is not clear how different variant combinations contribute to prolonged PFS in response to A+Ax treatment. However, examination of RNA expression in the DM tumors showed highly differential expression of the UTS2 gene, supporting a relationship between these genes and the existence of a unique phenotype in patients harboring these tumors (7).
In this study, the criteria for classifying a tumor as DM were further refined using a data-driven approach to create a “revised DM” (rDM) definition, reducing the number of patients from 170 with DM tumors to 139 with rDM tumors. Briefly, the inclusion of germline versus somatic variants in patients with mutations in the 10 genes of interest was evaluated by testing for association with PFS to determine whether there was a difference in the A+Ax treatment–by-biomarker interaction P value (see Methods). Following this analysis, the criteria for classifying variant types as rDM included only somatic mutations in 6 genes (ABCA1, CD163L1, DNMT1, MC1R, MYH7B, and STAB2) and only germline single-nucleotide polymorphisms in the other 4 genes (CROCC2/LOC728763, FOXO1, IL16 [P9L], and SPATA31C2). This refined classification further emphasized the PFS benefit in patients with rDM versus WT/S tumors in the A+Ax arm and highlighted a negative association with PFS in patients with rDM tumors in the sunitinib arm [A+Ax arm: HR, 2.42 (95% CI, 1.601–3.669); P < 0.0001; sunitinib arm: HR, 0.68 (95% CI, 0.499–0.934); P = 0.0162; Supplementary Fig. S4]. Using this refined definition of DM tumors, we examined our data sets for tumor tissue and peripheral markers with the goal of identifying differences between the mutational subgroups that might explain the unique phenotype of patients with rDM tumors and/or contribute to prolonged PFS in response to A+Ax. Previously reported TIL subset proportions in pretreatment tumor samples from patients in the A+Ax arm were examined to compare the rDM and WT/S subgroups (Fig. 3; ref. 7). In the rDM subgroup, but not the WT/S subgroup, longer PFS was observed in patients whose pretreatment tumors contained memory B cells and who had low levels of naïve B cells compared with the corresponding comparator subgroups. In contrast, PFS was longer in the WT/S subgroup in patients with high activated NK cells and low resting NK cells versus comparator subgroups. In both molecular subgroups, longer PFS was observed in patients with high levels of CD8+ T cells, and longer PFS was also seen in patients with rDM tumors who had high levels of activated antitumor (M1) macrophages and all patients who had low levels of resting-state (M0) macrophages, though these trends were less striking.
Figure 3.
PFS in the A+Ax arm according to mutation subgroup and proportion of different tumor-infiltrating lymphocyte subsets in pretreatment tumors. Two-sided P values comparing median subgroups within molecular subgroups were calculated using the log-rank test. aP values for treatment-by-biomarker interaction from a Cox model including treatment and biomarker, and the interaction term P value was smaller than 0.1 for naïve B cells (P = 0.0487), memory B cells (P = 0.0895), and M1 macrophages (P = 0.0717) using a 2-sided Wald test. bHazard ratios were calculated using the Cox proportional hazards model with < median used as the reference group, except for memory B cells, where the median value was 0 and 0/absence was used as the reference group. An HR of <1 indicates longer PFS in the ≥ median (or >0/presence) subgroup, whereas an HR of >1 indicates longer PFS in the < median (or 0/absence) subgroup. NE, not evaluable. PFS in the A+Ax arm according to mutation subgroup and proportion of different tumor-infiltrating lymphocyte subsets in pretreatment tumors. Two-sided P values comparing median subgroups within molecular subgroups were calculated using the log-rank test. aP values for treatmentby- biomarker interaction from a Cox model including treatment and biomarker, and the interaction term P value was smaller than 0.1 for naïve B cells (P = 0.0487), memory B cells (P = 0.0895), and M1 macrophages (P = 0.0717) using a 2-sided Wald test. bHazard ratios were calculated using the Cox proportional hazards model with < median used as the reference group, except for memory B cells, where the median value was 0 and 0/absence was used as the reference group. An HR of <1 indicates longer PFS in the ≥ median (or >0/presence) subgroup, whereas an HR of >1 indicates longer PFS in the < median (or 0/absence) subgroup. NE, not evaluable. PFS in the A+Ax arm according to mutation subgroup and proportion of different tumor-infiltrating lymphocyte subsets in pretreatment tumors. Two-sided P values comparing median subgroups within molecular subgroups were calculated using the log-rank test. aP values for treatmentby- biomarker interaction from a Cox model including treatment and biomarker, and the interaction term P value was smaller than 0.1 for naïve B cells (P = 0.0487), memory B cells (P = 0.0895), and M1 macrophages (P = 0.0717) using a 2-sided Wald test. bHazard ratios were calculated using the Cox proportional hazards model with < median used as the reference group, except for memory B cells, where the median value was 0 and 0/absence was used as the reference group. An HR of <1 indicates longer PFS in the ≥ median (or >0/presence) subgroup, whereas an HR of >1 indicates longer PFS in the < median (or 0/absence) subgroup. NE, not evaluable. PFS in the A+Ax arm according to mutation subgroup and proportion of different tumor-infiltrating lymphocyte subsets in pretreatment tumors. Two-sided P values comparing median subgroups within molecular subgroups were calculated using the log-rank test. aP values for treatmentby- biomarker interaction from a Cox model including treatment and biomarker, and the interaction term P value was smaller than 0.1 for naïve B cells (P = 0.0487), memory B cells (P = 0.0895), and M1 macrophages (P = 0.0717) using a 2-sided Wald test. bHazard ratios were calculated using the Cox proportional hazards model with < median used as the reference group, except for memory B cells, where the median value was 0 and 0/absence was used as the reference group. An HR of <1 indicates longer PFS in the ≥ median (or >0/presence) subgroup, whereas an HR of >1 indicates longer PFS in the < median (or 0/absence) subgroup. NE, not evaluable.
Proportions of TIL subsets in pretreatment tumors were compared between mutation subgroups (Table 2). Relative to WT/S tumors, rDM tumors had higher proportions of follicular helper T cells (P = 0.0006) and M1 macrophages (P = 0.0315) and numerically higher proportions of regulatory T cells and CD8+ T cells, as well as numerically lower proportions of resting memory CD4+ T cells. In comparisons of circulating cell populations at baseline between mutation subgroups, rDM tumors versus WT/S tumors had lower numbers of all populations, with marked differences for total leukocytes, monocytes, and neutrophils (P < 0.0001, P = 0.0019, and P = 0.0013, respectively).
Table 2.
Comparison of the median proportions of tumor-infiltrating leukocyte subsets, number of peripheral blood cell populations, and concentration of circulating proteins in pretreatment tumors by mutation subgroup.
| Infiltrating immune subpopulations (proportions of total TILs) | |||
|---|---|---|---|
| WT/S mutant tumors (n = 563), median (95% CI) |
rDM tumors (n = 124), median (95% CI) |
P valuea | |
| Follicular helper T cells | 0.045 (0.040–0.050) | 0.066 (0.049–0.076) | 0.0006 |
| M1 macrophages | 0.091 (0.085–0.098) | 0.106 (0.095–0.121) | 0.0315 |
| Resting memory CD4+ T cells | 0.218 (0.207–0.230) | 0.200 (0.180–0.222) | 0.0504 |
| T regulatory cells | 0.002 (0.001–0.004) | 0.004 (0.001–0.010) | 0.0536 |
| CD8+ T cells | 0.051 (0.043–0.061) | 0.061 (0.048–0.082) | 0.1276 |
| Circulating cell populations (×109/L) | |||
| WT/S mutant tumors (n = 601), median (95% CI) |
rDM tumors (n = 139), median (95% CI) |
P value | |
| Leukocytes | 7.100 (6.900–7.300) | 6.270 (6.000–6.700) | <0.0001 |
| Lymphocytes | 1.580 (1.500–1.610) | 1.520 (1.410–1.630) | 0.3224 |
| Monocytes | 0.600 (0.560–0.600) | 0.500 (0.440–0.540) | 0.0019 |
| Neutrophils | 4.480 (4.320–4.600) | 4.100 (3.740–4.400) | 0.0013 |
| Basophils | 0.040 (0.030–0.040) | 0.030 (0.020–0.040) | 0.3168 |
| Eosinophils | 0.160 (0.140–0.190) | 0.150 (0.110–0.200) | 0.9477 |
| Platelets | 259 (250–266) | 250 (230–278) | 0.6339 |
| Circulating protein concentrations | |||
| WT/S mutant tumors (n = 513), median (95% CI) |
rDM tumors (n = 132), median (95% CI) |
P value | |
| Eotaxin-1 (pg/mL) | 170.0 (153.0–182.0) | 142.5 (118.0–157.0) | 0.0095 |
| Factor VII (ng/mL) | 451.0 (436.0–468.0) | 471.5 (453.0–494.0) | 0.1131 |
| IL17b (pg/mL) | 2.6 (2.6–2.6)c | 2.6 (2.6–2.6) | 0.0674 |
| IL7b (pg/mL) | 51.0 (51.0–51.0) | 51.0 (51.0–51.0)c | 0.0034 |
| MCP-1 (pg/mL) | 394.0 (372.0–421.0) | 331.5 (306.0–386.0) | 0.0060 |
| MIP-1β (pg/mL) | 278.0 (268.0–302.0) | 333.5 (307.0–368.0) | 0.0035 |
| VEGF (pg/mL) | 299.0 (281.0–317.0) | 323.5 (284.0–357.0) | 0.0755 |
| CRPd (mg/L) | 7.8 (6.5–9.3) | 5.6 (4.0–10.7) | 0.0762 |
aTwo-sided P value is based on the Wilcoxon rank sum test; a statistical threshold value of P < 0.05 highlights observations of likely biological relevance.
bMedian = LLOQ.
cHigher mean value and greater proportion of patients > LLOQ.
dWT/S mutant subgroup (n = 574); rDM subgroup (n = 134).
CD4+, helper T cells; CD8+, killer T cells.
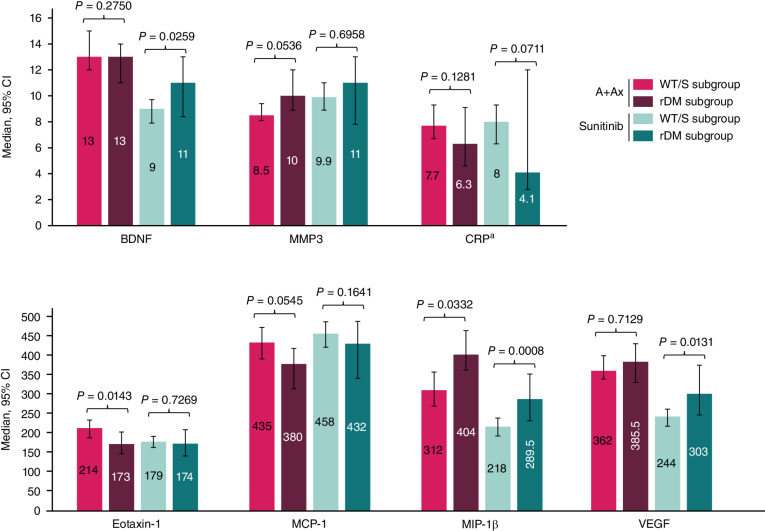
Circulating protein levels also differed at baseline between mutation subgroups (Table 2). Patients harboring rDM tumors showed lower concentrations of eotaxin-1 (P = 0.0095) and monocyte chemoattractant protein-1 (MCP-1; P = 0.0060) and higher macrophage inflammatory protein-1 beta (MIP-1β; P = 0.0035) and IL7 concentrations (P = 0.0034). Circulating protein levels were also compared between mutation subgroups on treatment (cycle 2 day 1; Fig. 4). In the A+Ax arm, the rDM subgroup had substantially lower eotaxin-1 (P = 0.0143), substantially higher MIP-1β concentrations (P = 0.0332), numerically higher matrix metalloproteinase-3 concentrations (MMP3), and lower MCP-1 concentrations. In the sunitinib arm, concentrations of MIP-1β, brain-derived neurotrophic factor (BDNF), and VEGF were markedly different between the rDM and WT/S subgroups (P = 0.0008, P = 0.0259, and P = 0.0131, respectively); a similar but smaller difference was observed for CRP concentration.
Figure 4.
Circulating cytokine and chemokine levels with the greatest differences at cycle 2 day 1 by mutation subgroup in the A+Ax or sunitinib arm. Median ± 95% CI for each analyte. A+Ax: WT/S mutant subgroup, n = 209; rDM subgroup, n = 66; sunitinib: WT/S mutant subgroup, n = 201; rDM subgroup, n = 46. aN values for CRP analysis: A+Ax: WT/S mutant subgroup, n = 246; rDM subgroup, n = 70; sunitinib: WT/S mutant subgroup, n = 245; rDM subgroup, n = 50. A statistical threshold value of P < 0.05 highlights observations of likely biological relevance.
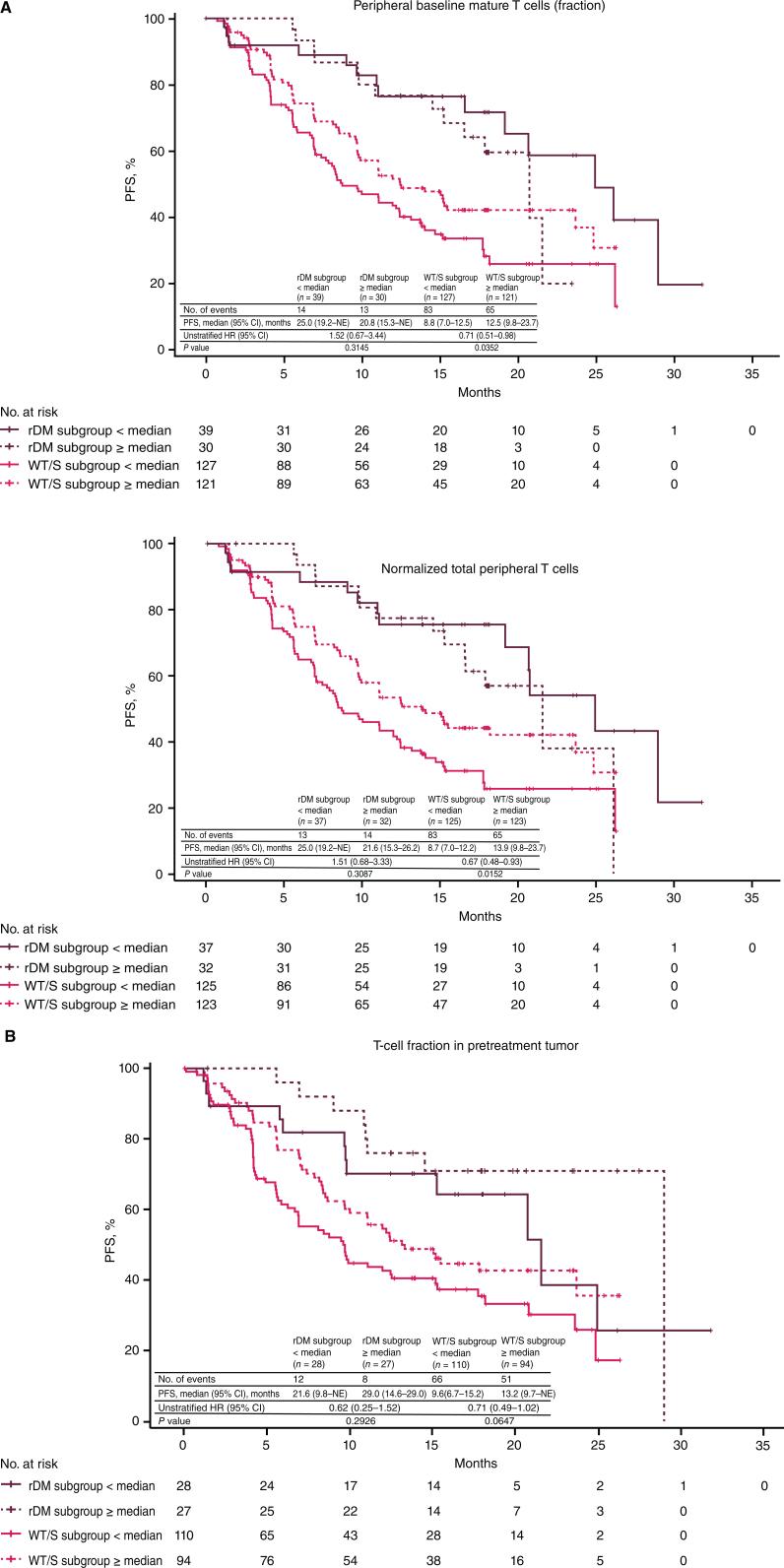
The only difference observed between mutation-defined molecular subgroups in TCR quantitation metrics was a numerical difference in the normalized number of total T cells in pretreatment tumors in the rDM versus WT/S subgroup [2.817 (interquartile range, 1.514–5.389) vs. 2.412 (interquartile range, 1.009–4.633), respectively; P = 0.0748; Supplementary Fig. S5]; all other comparisons of T-cell quantitation or repertoire measures at baseline or following treatment showed no differences between the molecular subgroups (Supplementary Fig. S6). Lastly, striking differences in PFS with A+Ax were seen in the WT/S subgroup for baseline TCR quantitation metrics, which were not evident when examined in the full treatment arm patient population (Fig. 5). Specifically, PFS with A+Ax was prolonged in patients with WT/S tumors with high baseline peripheral T-cell fractions (P = 0.0352) and normalized total T cells (P = 0.0152; Fig. 5A); a similar but smaller difference in PFS with high pretreatment tumor T-cell fraction was observed in the WT/S and rDM subgroups (Fig. 5B).
Figure 5.
PFS according to mutation subgroup in the A+Ax arm for T-cell quantitation metrics. A, PFS according to mutation subgroup in the A+Ax arm based on peripheral baseline T-cell fraction (top) and normalized total T cells (bottom). B, PFS according to mutation subgroup in the A+Ax arm based on T-cell fraction in pretreatment tumor samples. Hazard ratios were calculated using the Cox proportional hazards model with < median used as the reference group. Two-sided P values comparing median subgroups within molecular subgroups were calculated using the log-rank test; a statistical threshold value of P < 0.05 highlights observations of likely biological relevance.
DISCUSSION
Using one of the largest RCC data sets ever generated, exploratory analyses of peripheral blood and tumor characteristics in patients with aRCC in the JAVELIN Renal 101 trial provided novel insights into immunologic mechanisms associated with efficacy outcomes with 1L A+Ax or sunitinib treatment. Our findings support the previously described prognostic utility of some common blood-based biomarkers in aRCC, including levels of circulating cell populations (e.g., neutrophils and platelets) and peripheral markers of systemic inflammation (e.g., IL6 and CRP; refs. 13, 18–24). Additionally, some of our novel observations may prove to have prognostic or predictive significance, and although most were found to be treatment specific, their biological significance may extend beyond simple prognostic utility.
Comparisons of the A+Ax and sunitinib arms suggest potential differences in the extent and mechanisms of immunomodulation during treatment. In the A+Ax arm, baseline levels of circulating innate immune cell subsets, but not lymphocyte populations, were associated with PFS, contrary to expectations for an ICI-containing regimen. In the sunitinib arm, baseline levels of lymphocytes were positively associated with PFS; numbers and fractions of peripheral T cells were associated with prolonged PFS and increased with treatment, and oscillations with treatment were seen in various peripheral cell populations. These data support the proposed hypothesis that sunitinib has immunomodulatory effects (25, 26) that may be partially driven by inhibitory effects of sunitinib on targets other than the VEGF receptor, including platelet-derived growth factor receptor (PDGFR), KIT receptor tyrosine kinase, and fms-like tyrosine kinase 3 (FLT3), which are involved in the growth and survival of leukocytes (27–29).
However, we found that sunitinib treatment results in minimal modulation of the TCR repertoire; smaller changes in PFS were observed in patients with a lower degree of TCR repertoire modulation, whereas markedly longer PFS was observed in patients with greater numbers of T cells at baseline and during treatment. These results suggest that associations with the efficacy of sunitinib may be reflective of immunologic fitness (i.e., the potential to regain depleted immune cell populations during sunitinib treatment rather than activate new immune responses).
In contrast, the relatively minor changes in numbers of circulating cell subsets with A+Ax might be explained by the relatively high affinity of axitinib for VEGF receptors but not KIT or PDGFR (30), resulting in less inhibition of leukocyte survival and no association between leukocyte numbers and PFS. For lymphocytes and T cells in particular, the absence of any association with PFS in the A+Ax arm suggests two potential explanations. First, A+Ax treatment may elicit a focused, tumor-localized immune response that is difficult to distinguish within the peripheral lymphocyte and T-cell subpopulations. Second, the proposed innate immune mechanism of avelumab may contribute to treatment responses, diluting any statistical (but not biological) impact of cytotoxic T cells on PFS. The first hypothesis is supported by TCR analyses, in which smaller changes in T-cell quantitation metrics and a greater change in the TCR repertoire were observed with A+Ax versus sunitinib. This suggests that T cell–mediated immune responses are stimulated with A+Ax but may only involve a small population of tumor-reactive clones, as supported recently for melanoma (31), and/or predominantly occur in the tumor.
In support of the second hypothesis, recent analyses from the phase III JAVELIN Bladder 100 trial of avelumab 1L maintenance in advanced urothelial carcinoma suggest that NK cells may contribute to the destruction of tumor cells by recognizing avelumab-bound tumor cells via their FcγRIIa and FcγRIIIa receptors; this is consistent with findings of previous in vitro studies (8, 9, 32). In addition, in previous biomarker analyses from JAVELIN Renal 101, the 26-GES associated with PFS identified 6 genes associated with NK cell–mediated cytotoxicity (7), supporting a role for NK cells in the clinical activity of avelumab.
Analyses of the tumor molecular genotypes (7) revealed differences in baseline phenotypes in both the tumor and periphery in patients in the rDM subgroup, as well as distinct treatment-emergent changes in peripheral biomarker levels. In analyses of baseline peripheral biomarkers, differences between the mutation-defined subgroups were seen in levels of innate immune cytokines, chemokines, and cell populations. In contrast, none of the peripheral measures of adaptive immune cell populations showed differences between the molecular subgroups. However, the rDM subgroup had more T cells in tumors at baseline, and PFS with A+Ax was numerically longer in patients with high versus low levels of baseline CD8+ T cells in tumors regardless of molecular subgroup; this suggests that these cells are involved in treatment responses in both mutation-defined subgroups. Interestingly, only the WT/S subgroup showed associations of PFS with a higher tumor T-cell fraction and higher peripheral T-cell quantitation metrics; these associations were not seen in the A+Ax arm as a whole. Overall, these findings confirm the role of the canonical cytotoxic T cell–mediated mechanism of action presumed to be induced by ICIs (33) and suggest that this mechanism may play a larger role in treatment response in patients with WT/S versus rDM tumors.
TIL deconvolution analyses also suggested that T cell–independent mechanisms in response to A+Ax differed between mutation-defined subgroups. In the WT/S subgroup, PFS with A+Ax was longer in patients with high levels of activated NK cells and low levels of resting NK cells versus comparator subgroups, further supporting NK cell–mediated ADCC with avelumab. However, in the rDM subgroup, prolonged PFS with A+Ax was associated with low levels of naïve B cells and the presence of memory B cells and trended with high levels of M1 macrophages, suggesting a potential role for antibody-dependent cellular phagocytosis (ADCP; ref. 34) rather than ADCC. ADCP could be triggered or enhanced by treatment with A+Ax via a direct mechanism [opsonization of tumor cells expressing PD-L1 by avelumab binding (35)] and/or an indirect mechanism [relief of PD-L1–mediated inhibition of follicular helper T cells (Tfh) via checkpoint blockade with avelumab, facilitating increased antitumor antibody production by infiltrating B cells (36)]. Recent work has described the likely importance of these interactions in tertiary lymphoid structures within tumors and the potential effects of ICI therapy on their component cell types (37, 38).
In rDM tumors, ADCP could also be facilitated by the observed higher levels of TIL recruitment and migration into the tumor, including the higher proportions of M1 macrophages and Tfh cells present in these tumors as compared with WT/S tumors. Further, patients with rDM tumors had lower MCP-1 and higher MIP-1β levels at baseline and following treatment with A+Ax versus patients with WT/S tumors, which may translate to less monocyte chemotaxis and fewer immunosuppressive tumor-associated macrophages in the tumor microenvironment (39). MIP-1β production by monocytes or macrophages is induced by IL7 (40), and IL7 has been shown to drive M1 macrophage differentiation in a mouse model (41). IL7 was detectable at baseline in a greater proportion of patients with rDM tumors versus WT/S tumors. These phenotypic observations provide a potential mechanism for the prolonged PFS observed in patients with rDM tumors in the A+Ax arm and represent an area of possible future study within a biomarker-defined pan-tumor population.
To better understand the potential driver(s) of this phenotype, we revisited the previously described differential RNA expression profiles of DM versus WT/S tumors (7). We previously reported that UTS2, which encodes urotensin II (UII), is expressed at higher levels in DM pretreatment tumor tissue and that higher UTS2 levels were associated with prolonged PFS in all patients treated with A+Ax, irrespective of mutant subgroup (7). UII is a proinflammatory cytokine/chemokine and a potent regulator of vasoconstriction; through these mechanisms, UII may potentially contribute to increased vascular permeability because of inflammation-mediated vessel damage (42). This may provide a mechanism for the increased infiltration observed in rDM tumors. Further, studies performed in UII knockout mice compared with WT mice showed a differential cholesterol synthesis in hepatocytes and macrophages, supporting a role for UII signaling in macrophage function (43). Lastly, as a chemokine known to signal through the mitogen-activated protein kinase, interferon, and nuclear factor kappa B pathways (42), UII may contribute to both the antiangiogenic and immunomodulatory mechanisms of A+Ax. Therefore, the observed rDM phenotype of increased tumor infiltration and differential cytokine/chemokine profiles may be driven in part by higher levels of UII, which suggests that high UII expression could be used as a potential biomarker to identify patients who may benefit from A+Ax treatment regardless of tumor biology, including patients with WT/S tumors. Irrespective of the mechanism(s), our data suggest that an M1-favorable tumor microenvironment, the antitumor activity of Tfh cells, B cells, and M1 macrophages, and canonical cytotoxic CD8+ T-cell response to ICIs may contribute to the extended PFS benefit seen in patients with rDM tumors treated with A+Ax. As noted previously, the clinical application of these observations may extend beyond aRCC (7); multiple other tumor types in The Cancer Genome Atlas exhibited variants in the 10 genes of interest at frequencies ranging from ∼5%–25%, suggesting that A+Ax treatment in other tumor types may provide similar benefit in patients with rDM tumors. However, additional work is required to confirm these hypotheses.
In conclusion, dual analyses of baseline and longitudinal peripheral biomarkers and baseline tumor molecular data from the JAVELIN Renal 101 trial provide evidence of different immunomodulatory mechanisms in patients with aRCC treated with A+Ax versus sunitinib. Our findings further suggest that the adaptive immunologic activity of A+Ax may be restricted to a subset of responsive tumor-reactive T-cell clones. We also found evidence for a distinct phenotype and biology for the rDM tumor subgroup, which helped elucidate a role for the innate immune system in the mechanism of action of A+Ax treatment in aRCC via antibody-dependent processes that may differ in the molecular subgroups. Overall, these integrated findings demonstrate the utility of longitudinal studies in understanding the response of both the tumor and the immune system to treatment with different regimens and suggest that although cytotoxic T cells are involved in response to A+Ax treatment in patients with aRCC, the overall mechanism of action is more complex and dependent on patient and tumor characteristics and the treatment given.
METHODS
Trial Design and Patient Population
The design of the phase III JAVELIN Renal 101 trial (trial registration ID: NCT02684006) has been described in detail previously, and results are reported per CONSORT study guidelines (1, 2). Briefly, the trial enrolled adults (N = 886) with previously untreated aRCC with a clear-cell component. Randomization (1:1) to A+Ax or sunitinib was stratified by Eastern Cooperative Oncology Group performance status (0 vs. 1) and geographic region (United States vs. Canada and Western Europe vs. rest of the world). Avelumab was administered intravenously at a dose of 10 mg/kg every 2 weeks. Axitinib was administered orally at a dose of 5 mg twice daily. Sunitinib was administered orally at a dose of 50 mg once daily for 4 weeks in a 6-week cycle; for alignment within the study, a single cycle consisted of 6 weeks for both treatment arms. The trial was conducted in accordance with the ethics principles of the Declaration of Helsinki and the Good Clinical Practice guidelines, defined by the International Council for Harmonization. All patients provided written informed consent.
Peripheral Blood and Tumor Analyses
Blood samples were collected during screening and on multiple days within cycles 1, 2, and 3 (prior to treatment), including collection of whole blood in dipotassium ethylenediaminetetraacetic acid (K2 EDTA) optimized for DNA analysis and collection of blood in K2 EDTA or silica-coated serum tubes for plasma and serum separation, respectively. The total number of patients and samples available for each analysis was dependent on patient consent, local regulations, paired sample availability, volume limitations, and/or local laboratory data reporting by clinical sites.
Circulating peripheral cell population counts (as 109/L) were collected from site-reported results for local laboratory complete blood count with differential testing. Similarly, CRP data were collected from site-reported results for local CRP testing. For analyses of peripheral cell population counts and CRP, low values were defined as < median across all evaluable patients, and high values were defined as ≥ median.
Cytokine and chemokine concentrations in patient serum were determined using 2 multiplexed panels that assayed 32 total analytes (Rules-Based Medicine); analytes included BDNF, eotaxin-1, factor VII, granulocyte–macrophage colony-stimulating factor (GM-CSF), intercellular adhesion molecule 1 (ICAM-1), interferon-gamma (IFNγ), IL1α, IL1β, IL1Ra, IL2, IL3, IL4, IL5, IL6, IL7, IL8, IL10, IL12p40, IL12p70, IL15, IL17, IL18, IL23, MIP-1α, MIP-1β, MMP3, MMP9, MCP-1, stem cell factor (SCF), tumor necrosis factor alpha (TNFα), TNF beta (TNFβ), and VEGF. Briefly, multiplexed cocktails of biotinylated reporter antibodies were added to the sample and developed using an excess of streptavidin-phycoerythrin solution, which was then analyzed on a Luminex instrument to provide a quantitative readout of concentration (pg/mL or ng/mL; units vary by analyte). To minimize technical variability, all serum specimens analyzed were batch tested with the same reagent lots, and paired patient specimens were analyzed on the same plates. Of the 32 proteins analyzed, only cytokines and chemokines with levels above the lower limit of quantitation (LLOQ) for a significant fraction of patients who also showed a trend or association with response are shown here. For cytokines and chemokine analytes that were analyzed, the LLOQ value was substituted for a reported assay result of “<[LLOQ],” and low values were defined as ≤ median of all patients (which may be equal to the LLOQ for a given analyte); high values were defined as > median.
TCR-seq was performed by Adaptive Biotechnologies. For the peripheral repertoire, genomic DNA (gDNA) was extracted from frozen K2 EDTA whole blood specimens by Adaptive Biotechnologies, and sequencing of the TCRB-CDR3 region was performed at deep resolution. For the pretreatment tumor TCR repertoire, remnant gDNA from the same pool previously used to perform whole-exome sequencing (7) was provided to Adaptive Biotechnologies and used to sequence the TCRB-CDR3 region at survey depth. Pairwise analyses were performed by Adaptive Biotechnologies and transferred to the sponsor for all possible specimen pairs for a given patient, including blood–blood and tumor–blood pairs (see Statistical Analysis).
Mutations in pretreatment tumor tissue were determined using whole-exome sequencing, and TIL deconvolution was performed using whole-transcriptome sequencing data as reported previously (7). The criteria for classifying a tumor as DM were refined using a data-driven approach, building on previously described observations regarding germline versus somatic variants in patients with mutations in the 10 genes of interest (7). Inclusion of germline versus somatic variants for each gene was evaluated by testing for association with PFS to determine whether there was a difference in the treatment-by-biomarker interaction P value; a Cox proportional hazards model was used, with the treatment arm and mutation status as independent variables. Four genes of interest were associated with prolonged PFS in the A+Ax arm for germline variants but not for somatic variants (CROCC2/LOC728763, FOXO1, IL16 [P9L], and SPATA31C2). The criteria for classifying a tumor as DM were therefore updated to include only germline variants (i.e., single-nucleotide polymorphisms) for these 4 genes and somatic variants for the other 6 genes of interest (ABCA1, CD163L1, DNMT1, MC1R, MYH7B, and STAB2), resulting in a revised DM (rDM) population with a reduction in the total number of qualifying tumors from 170 to 139 (7).
Statistical Analysis
All efficacy data reported in this article are based on the second interim analysis of the JAVELIN Renal 101 trial (data cutoff, January 2019). Kaplan–Meier estimates of PFS based on investigator assessment per Response Evaluation Criteria in Solid Tumors version 1.1 (44) were presented by treatment and biomarker subgroups together with a summary of the median PFS time with 2-sided 95% CIs (45). HRs and 95% CIs were calculated using the Cox proportional hazards model with and without adjustment for age and sex with no observable differences; therefore, nonadjusted data are reported. Similarly, PFS analyses using biomarker subgroup defined by median split were conducted with both the global medians for each time point as well as treatment arm–specific medians for on-study time points (cycle 2 day 1) with no changes in interpretation; therefore, data derived using global medians are reported. For comparisons of PFS between subgroups defined by different characteristics, 2-sided P values were calculated using log-rank tests. To assess treatment-by-biomarker interactions in subgroups, 2-sided P values were calculated using the Cox proportional hazards model with the treatment arm and the relevant median value in the overall population for each characteristic as independent variables. Because the JAVELIN Renal 101 trial was not designed a priori to evaluate the various biomarker questions under investigation, the exploratory analyses reported are subject to both type I and type II errors because of multiple testing and lack of statistical power, respectively. Because the analyses were intended to be descriptive and hypothesis-generating, the P values are presented without correction for multiple comparisons to reduce potential type II error. Analytes and comparisons of interest were identified and prioritized for further investigation using a statistical threshold of/near <0.05 to highlight observations of likely biological relevance. These analyses should be interpreted alongside other factors including sample size, effect size, and the exploratory nature of the analyses while also acknowledging the limitation of using P values as the evidence against the null hypothesis (46–48).
For TCR-seq, total T-cell numbers were normalized to the nanograms of input gDNA for that specimen to control for differential amounts of available gDNA between specimens. The normalized number of total T-cell clones in a given specimen was extrapolated using the number of clones shared between paired patient blood specimens and the fraction of those clones that were shared in the specimen from the time point of interest; the number was then normalized to the input gDNA. Only productively rearranged TCR clones with a combined total of 5 reads between the paired specimens are included in the count of shared clones in pairwise analyses. Log2 fold changes are calculated as log2 (ratio of later time point to earlier time point). To analyze the TCR repertoire, the Morisita index, beta-binomial model, and Simpson clonality were assessed (49–51). The Morisita index calculates the similarity between 2 repertoires, giving a value between 0 (unrelated) and 1 (identical; ref. 49). Maximum clone expansion and contraction represent the greatest observed fold increase and decrease, respectively, in clone frequency at the later time point. The beta-binomial model accounts for normal variation levels in clone frequencies observed in the absence of immune responses and reports only the number of clones whose frequencies are likely to have changed as part of repertoire modulation (50). Simpson clonality (i.e., square root of 1 – Simpson's Diversity index) describes the shape of T-cell clone distribution, with values approaching 1 indicating a more oligoclonal population (49, 51). For comparisons of T-cell metrics between treatment arms or cell population or circulating protein concentrations in mutation-defined subgroups, P values were calculated using the Wilcoxon rank sum test.
Data Availability Statement
Upon request, and subject to review, Pfizer will provide the data that support the findings of this study. Subject to certain criteria, conditions, and exceptions, Pfizer may also provide access to the related individual deidentified participant data. See https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information.
Supplementary Material
Supplementary Figure S1 shows forest plots of PFS according to numbers of circulating cell populations in the overall trial population (both arms combined) at baseline. Supplementary Figure S2 shows forest plots of PFS with A+Ax or sunitinib according to cytokine, chemokine, or protein concentrations in peripheral blood at baseline and during treatment (cycle 2 day 1). Supplementary Figure S3 shows PFS according to metrics describing TCR repertoire change from baseline to cycle 2 day 1 with A+Ax vs sunitinib. Supplementary Figure S4 shows PFS according to updated mutation subgroup definitions and treatment group. Supplementary Figure S5 shows normalized numbers of total T cells in pretreatment tumors according to molecular subgroup. Supplementary Figure S6 shows peripheral T-cell receptor quantitation and clonality metrics at baseline and cycle 2 day 1 by mutation subgroup.
Acknowledgments
The authors would like to thank the patients and their families, investigators, coinvestigators, and the study teams at each of the participating centers, as well as Keith Ching of Pfizer Computational Biology and Shibing Deng of Pfizer Biostatistics for data analysis. This trial was sponsored by Pfizer and was previously conducted under an alliance between the healthcare business of Merck KgaA, Darmstadt, Germany (CrossRef Funder ID: 10.13039/100009945) and Pfizer. Medical writing support was provided by Katherine Quiroz-Figueroa, PhD, of Nucleus Global and funded by the healthcare business of Merck KGaA, Darmstadt, Germany and Pfizer. Patients treated at Memorial Sloan Kettering Cancer Center were supported in part by the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748).
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T.K. Choueiri reports nonfinancial support from the healthcare business of Merck KGaA, Darmstadt, Germany, and nonfinancial support from Pfizer during the conduct of the study; grants and personal fees from Alkermes, Aravive, AstraZeneca, Aveo Oncology, Bayer, Bristol Myers Squibb, Calithera, Circle Pharma, CME events (Peerview, OncLive, MJH, and others), Eisai, Exelixis, GSK, IQVA, Infinity, Ipsen, Jansen, Kanaph, Lilly, Merck & Co., Kenilworth, NJ, Nikang Therapeutics, Novartis, Nuscan, Pfizer, Roche, Sanofi-Aventis, Surface Oncology, Takeda, Tempest, the healthcare business of Merck KGaA, Darmstadt, Germany, and Up-To-Date outside the submitted work; reports institutional patents filed on molecular alterations and immunotherapy response/toxicity and ctDNA; reports equity for Osel, Pionyr Immunotherapeutics, Precede Biosciences, and Tempest Therapeutics; has served on committees for ACCRU, ASCO/ESMO, GU Steering Committee, KidneyCAN, and NCCN; reports that medical writing and editorial assistance support may have been funded by communications companies in part; reports no speakers bureau; has mentored several non-US citizens on research projects with potential funding (in part) from non-US sources and foreign components; reports that the institution (Dana-Farber Cancer Institute) may have received additional independent funding of drug companies and/or royalties potentially involved in research around the subject matter; and is supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE (2P50CA101942-16) and Program 5P30CA006516-56, the Kohlberg Chair at Harvard Medical School and the Trust Family, Michael Brigham, Pan-Mass Challenge, and Loker Pinard Funds for Kidney Cancer Research at DFCI. A.C. Donahue reports personal fees from Pfizer, Instil Bio, Ideaya Biosciences, and St. John's Cancer Institute outside the submitted work. D.A. Braun reports grants and personal fees from Exelixis, personal fees from ScholarRock, Pfizer, Link Cell Therapeutics, Compugen, Accolade 2nd MD, DLA Piper, Merck & Co., Kenilworth, NJ, Elephas, Targeted Oncology, and AbbVie, and nonfinancial support from Curious and Fortress Biotech outside the submitted work. B.I. Rini reports research funding from Dragonfly Therapeutics, HiberCell, Incyte, Stata Oncology, ADC Therapeutics, Dracen Pharmaceuticals, Janssen, Adela, AstraZeneca, Pionyr, Tempus, VasGene Therapeutics, Gilead, POINT Biopharma, Bristol Myers Squibb, Pfizer, Daiichi Sankyo, Genentech, Arrowhead Pharmaceuticals, Exelixis, Surface Oncology, Aravive; and consulting for Bristol Myers Squibb, Pfizer, GNE/Roche, Aveo, Synthorx, the healthcare business of Merck KGaA, Darmstadt, Germany, Corvus, Surface Oncology, Aravive, Alkermes, Arrowhead, Eisai, Nikang Therapeutics, EUSA, Athenex, Debiopharm, HiberCell. T. Powles reports personal fees from AstraZeneca, Bristol Myers Squibb, Exelixis, Incyte, Ipsen, Merck & Co., Kenilworth, NJ, Novartis, Pfizer, Seagen, the healthcare business of Merck KGaA, Darmstadt, Germany, Astellas, Johnson & Johnson, Eisai, and Roche during the conduct of the study. J.B.A.G. Haanen reports grants from Asher Bio, BioNTech, Bristol Myers Squibb, Novartis, and Sastra Cell Therapy, other support from AZ, BioNTech, Bristol Myers Squibb, Iovance Bio, Instil Bio, Immunocore, Merck & Co., Kenilworth, NJ, Novartis, Molecular Partners, Roche, Sanofi, and T-Knife, and personal fees and other support from CureVac, Neogene Tx, Sastra Cell Therapy, and TRV outside the submitted work. J. Larkin has received honoraria from Bristol Myers Squibb, CRUK, Dynavax, Eisai, GSK, Incyte, iOnctura, Merck & Co., Kenilworth, NJ, Novartis, Pfizer, Roche, touchExperts, and touchIME; has provided consulting or advisory roles for Apple Tree, Boston Biomedical, Bristol Myers Squibb, GSK, Immunocore, Incyte, iOnctura, Iovance, Novartis, Pfizer, and YKT Global; has received speaker fees from Aptitude, AstraZeneca, Bristol Myers Squibb, Calithera, Eisai, Ervaxx, EUSA Pharma, GSK, Incyte, Ipsen, Merck & Co., Kenilworth, NJ, Novartis, Pierre Fabre, Pfizer, Roche, Seagen, and Ultimovacs; and received institutional research funding from Achilles Therapeutics, Aveo, Bristol Myers Squibb, Covance, Immunocore, Merck & Co., Kenilworth, NJ, Nektar Therapeutics, Novartis, Pfizer, Pharmacyclics, and Roche; and has received grants from Achilles Therapeutics, Aveo, Bristol Myers Squibb, Immunocore, Merck & Co., Kenilworth, NJ, Nektar Therapeutics, Novartis, Pfizer, Pharmacyclics, and Roche. X.J. Mu is an employee of Pfizer and reports other support from Pfizer during the conduct of the study and other support from Pfizer outside the submitted work; in addition, X.J. Mu has a patent for Pfizer issued. Jie Pu is an employee of Pfizer. R.E. Teresi is an employee of Pfizer and reports personal fees from Pfizer outside the submitted work. A. di Pietro is an employee of Pfizer and reports other support from Pfizer during the conduct of the study. P.B. Robbins was an employee of Pfizer and reports other support from Pfizer during the conduct of the study. R.J. Motzer reports grants from Pfizer; personal fees from Pfizer and the healthcare business of Merck KGaA, Darmstadt, Germany, during the conduct of the study; personal fees from Genentech/Roche, the healthcare business of Merck KGaA, Darmstadt, Germany, Eisai, and Exelixis; and grants from Genentech/Roche, the healthcare business of Merck KGaA, Darmstadt, Germany, Eisai, Exelixis, Takeda, Aveo, and Aveo outside the submitted work.
Authors’ Contributions
T.K. Choueiri: Conceptualization, data curation, supervision, investigation, writing–original draft, writing–review and editing. A.C. Donahue: Conceptualization, data curation, formal analysis, methodology, visualization, investigation, writing–original draft, writing–review and editing.D.A. Braun: Conceptualization, data curation, investigation, writing–review and editing. B.I. Rini: Data curation, investigation, writing–review and editing. T. Powles: Data curation, investigation, writing–review and editing. J.B.A.G. Haanen: Data curation, investigation, writing–review and editing. J. Larkin: Data curation, investigation, writing–review and editing. X.J. Mu: Data curation, software, formal analysis, validation, visualization, methodology, writing–review and editing. J. Pu: Software, formal analysis, validation, visualization, methodology, writing–review and editing. R.E. Teresi: Supervision, writing–review and editing. A. di Pietro: Conceptualization, supervision, writing–review and editing. P.B. Robbins: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. R.J. Motzer: Data curation, investigation, writing–review and editing.
References
- 1. Choueiri TK, Motzer RJ, Rini BI, Haanen J, Campbell MT, Venugopal B, et al. Updated efficacy results from the JAVELIN Renal 101 trial: first-line avelumab plus axitinib versus sunitinib in patients with advanced renal cell carcinoma. Ann Oncol 2020;31:1030–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MT, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bavencio (avelumab). Prescribing information. EMD Serono Rockland, MA, USA 2023. [Cited September 22, 2023]. Available from: https://www.emdserono.com/us-en/pi/bavencio-pi.pdf.
- 4. Inlyta (axitinib). Prescribing information. Pfizer 2022. [Cited September 22, 2023]. Available from: https://labeling.pfizer.com/showlabeling.aspx?id=759.
- 5. Bavencio (avelumab). Summary of product characteristics. Merck Europe B.V, Amsterdam, the Netherlands, an affiliate of the healthcare business of Merck KGaA, Darmstadt, Germany. 2023. [Cited September 22, 2023]. Available from: https://www.ema.europa.eu/en/documents/product-information/bavencio-epar-product-information_en.pdf.
- 6. Inlyta. Summary of Product Characteristics. 2021. [Cited September 22, 2023]. Available from: https://www.ema.europa.eu/en/documents/product-information/inlyta-epar-product-information_en.pdf.
- 7. Motzer RJ, Robbins PB, Powles T, Albiges L, Haanen JB, Larkin J, et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nat Med 2020;26:1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyerinas B, Jochems C, Fantini M, Heery CR, Gulley JL, Tsang KY, et al. Antibody-dependent cellular cytotoxicity activity of a novel anti-PD-L1 antibody avelumab (MSB0010718C) on human tumor cells. Cancer Immunol Res 2015;3:1148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Powles T, Sridhar SS, Loriot Y, Bellmunt J, Mu XJ, Ching KA, et al. Avelumab maintenance in advanced urothelial carcinoma: biomarker analysis of the phase 3 JAVELIN Bladder 100 trial. Nat Med 2021;27:2200–11. [DOI] [PubMed] [Google Scholar]
- 10. Choueiri TK, Larkin J, Pal S, Motzer RJ, Rini BI, Venugopal B, et al. Efficacy and correlative analyses of avelumab plus axitinib versus sunitinib in sarcomatoid renal cell carcinoma: post hoc analysis of a randomized clinical trial. ESMO Open 2021;6:100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu L, Li H, Gao Y, Ma X, Chen L, Li X, et al. The association of platelet count with clinicopathological significance and prognosis in renal cell carcinoma: a systematic review and meta-analysis. PLoS One 2015;10:e0125538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen J, Chen Z, Fan M, Lu H, Zhuang Q, He X. Prognostic value of pretreatment neutrophil count in metastatic renal cell carcinoma: a systematic review and meta-analysis. Cancer Manag Res 2019;11:5365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bilen MA, Rini BI, Voss MH, Larkin J, Haanen JBAG, Albiges L, et al. Association of neutrophil-to-lymphocyte ratio with efficacy of first-line avelumab plus axitinib vs. sunitinib in patients with advanced renal cell carcinoma enrolled in the phase 3 JAVELIN Renal 101 trial. Clin Cancer Res 2021;28:738–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Brian D, Prunty M, Hill A, Shoag J. The role of C-reactive protein in kidney, bladder, and prostate cancers. Front Immunol 2021;12:721989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanmamed MF, Carranza-Rua O, Alfaro C, Oñate C, Martín-Algarra S, Perez G, et al. Serum interleukin-8 reflects tumor burden and treatment response across malignancies of multiple tissue origins. Clin Cancer Res 2014;20:5697–707. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Zhang Y. Prognostic role of interleukin-6 in renal cell carcinoma: a meta-analysis. Clin Transl Oncol 2020;22:835–43. [DOI] [PubMed] [Google Scholar]
- 17. Zhao S, Wu D, Wu P, Wang Z, Huang J. Serum IL-10 predicts worse outcome in cancer patients: a meta-analysis. PLoS One 2015;10:e0139598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishihara H, Takagi T, Kondo T, Fukuda H, Tachibana H, Yoshida K, et al. Predictive impact of an early change in serum C-reactive protein levels in nivolumab therapy for metastatic renal cell carcinoma. Urol Oncol 2020;38:526–32. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki K, Terakawa T, Furukawa J, Harada K, Hinata N, Nakano Y, et al. C-reactive protein and the neutrophil-to-lymphocyte ratio are prognostic biomarkers in metastatic renal cell carcinoma patients treated with nivolumab. J Clin Oncol 2020;25:135–44. [DOI] [PubMed] [Google Scholar]
- 20. Pilskog M, Nilsen GH, Beisland C, Straume O. Elevated plasma interleukin 6 predicts poor response in patients treated with sunitinib for metastatic clear cell renal cell carcinoma. Cancer Treat Res Commun 2019;19:100127. [DOI] [PubMed] [Google Scholar]
- 21. Esteban E, Exposito F, Crespo G, Lambea J, Pinto A, Puente J, et al. Circulating levels of the Interferon-γ-regulated chemokines CXCL10/CXCL11, IL-6 and HGF predict outcome in metastatic renal cell carcinoma patients treated with antiangiogenic therapy. Cancers 2021;13:2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lalani AA, Xie W, Martini DJ, Steinharter JA, Norton CK, Krajewski KM, et al. Change in neutrophil-to-lymphocyte ratio (NLR) in response to immune checkpoint blockade for metastatic renal cell carcinoma. J Immunother Cancer 2018;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noguchi G, Nakaigawa N, Umemoto S, Kobayashi K, Shibata Y, Tsutsumi S, et al. C-reactive protein at 1 month after treatment of nivolumab as a predictive marker of efficacy in advanced renal cell carcinoma. Cancer Chemother Pharmacol 2020;86:75–85. [DOI] [PubMed] [Google Scholar]
- 24. Tomita Y, Larkin J, Venugopal B, Haanen J, Kanayama H, Eto M, et al. Association of C-reactive protein with efficacy of avelumab plus axitinib in advanced renal cell carcinoma: long-term follow-up results from JAVELIN Renal 101. ESMO Open 2022;7. 10.1016/j.esmoop.2022.100564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nuti M, Zizzari I, Napoletano C, Botticelli A, Calabro F, Gelibter A, et al. Immunomodulatory effects of tyrosine kinase inhibitors (TKIs) in renal cell carcinoma (RCC) patients. J Clin Oncol 35, 2017. (suppl 15; abstr e14506). [Google Scholar]
- 26. Mizuno R, Kimura G, Fukasawa S, Ueda T, Kondo T, Hara H, et al. Angiogenic, inflammatory and immunologic markers in predicting response to sunitinib in metastatic renal cell carcinoma. Cancer Sci 2017;108:1858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andrae N, Kirches E, Hartig R, Haase D, Keilhoff G, Kalinski T, et al. Sunitinib targets PDGF-receptor and Flt3 and reduces survival and migration of human meningioma cells. Eur J Cancer 2012;48:1831–41. [DOI] [PubMed] [Google Scholar]
- 28. Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci U S A 2009;106:1542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Papaetis GS, Syrigos KN. Sunitinib. BioDrugs 2009;23:377–89. [DOI] [PubMed] [Google Scholar]
- 30. Carmichael C, Lau C, Josephson DY, Pal SK. Comprehensive overview of axitinib development in solid malignancies: focus on metastatic renal cell carcinoma. Clin Adv Hematol Oncol 2012;10:307–14. [PubMed] [Google Scholar]
- 31. Puig-Saus C, Sennino B, Peng S, Wang CL, Pan Z, Yuen B, et al. Neoantigen-targeted CD8+ T cell responses with PD-1 blockade therapy. Nature 2023;615:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juliá EP, Amante A, Pampena MB, Mordoh J, Levy EM. Avelumab, an IgG1 anti-PD-L1 immune checkpoint inhibitor, triggers NK cell-mediated cytotoxicity and cytokine production against triple negative breast cancer cells. Front Immunol 2018;9:2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Granier C, De Guillebon E, Blanc C, Roussel H, Badoual C, Colin E, et al. Mechanisms of action and rationale for the use of checkpoint inhibitors in cancer. ESMO Open 2017;2:e000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weiskopf K, Weissman IL. Macrophages are critical effectors of antibody therapies for cancer. MAbs 2015;7:303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bakema JE, van Egmond M. Fc receptor-dependent mechanisms of monoclonal antibody therapy of cancer. Curr Top Microbiol Immunol 2014;382:373–92. [DOI] [PubMed] [Google Scholar]
- 36. Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia-Recio S, Mott KR, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell 2019;179:1191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang E, Ding C, Li S, Zhou X, Aikemu B, Fan X, et al. Roles and mechanisms of tumour-infiltrating B cells in human cancer: a new force in immunotherapy. Biomark Res 2023;11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garaud S, Dieu-Nosjean MC, Willard-Gallo K. T follicular helper and B cell crosstalk in tertiary lymphoid structures and cancer immunotherapy. Nat Commun 2022;13:2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoshimura T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: a foe or ally? Cell Mol Immunol 2018;15:335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Damås JK, Wæhre T, Yndestad A, Otterdal K, Hognestad A, Solum NO, et al. Interleukin-7-mediated inflammation in unstable angina. Circulation 2003;107:2670–6. [DOI] [PubMed] [Google Scholar]
- 41. Chen Z, Kim S-jChamberlain ND, Pickens SR, Volin MV, Volkov S, et al. The novel role of IL-7 ligation to IL-7 receptor in myeloid cells of rheumatoid arthritis and collagen-induced arthritis. J Immunol 2013;190:5256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun SL, Liu LM. Urotensin II: an inflammatory cytokine. J Endocrinol 2019;240:R107–17. [DOI] [PubMed] [Google Scholar]
- 43. Kiss RS, You Z, Genest J Jr, Behm DJ, Giaid A. Urotensin II differentially regulates macrophage and hepatic cholesterol homeostasis. Peptides 2011;32:956–63. [DOI] [PubMed] [Google Scholar]
- 44. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 45. Brookmeyer R, Crowley J. A confidence interval for the median survival time. Biometrics 1982;38:29–41. [Google Scholar]
- 46. Wasserstein RL, Lazar NA. The ASA statement on p-values: context, process, and purpose. Am Stat 2016;70:129–33. [Google Scholar]
- 47. Betensky RA. The p-value requires context, not a threshold. Am Stat 2019;73( sup1):115–7. [Google Scholar]
- 48. Wasserstein RL, Schirm AL, Lazar NA. Moving to a world beyond “p < 0.05.” Am Stat 2019;73( sup1):1–19. [Google Scholar]
- 49. Chiffelle J, Genolet R, Perez MAS, Coukos G, Zoete V, Harari A. T-cell repertoire analysis and metrics of diversity and clonality. Curr Opin Biotechnol 2020;65:284–95. [DOI] [PubMed] [Google Scholar]
- 50. Rytlewski J, Deng S, Xie T, Davis C, Robins H, Yusko E, et al. Model to improve specificity for identification of clinically-relevant expanded T cells in peripheral blood. PLoS One 2019;14:e0213684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simpson EH. Measurement of diversity. Nature 1949;163:688. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 shows forest plots of PFS according to numbers of circulating cell populations in the overall trial population (both arms combined) at baseline. Supplementary Figure S2 shows forest plots of PFS with A+Ax or sunitinib according to cytokine, chemokine, or protein concentrations in peripheral blood at baseline and during treatment (cycle 2 day 1). Supplementary Figure S3 shows PFS according to metrics describing TCR repertoire change from baseline to cycle 2 day 1 with A+Ax vs sunitinib. Supplementary Figure S4 shows PFS according to updated mutation subgroup definitions and treatment group. Supplementary Figure S5 shows normalized numbers of total T cells in pretreatment tumors according to molecular subgroup. Supplementary Figure S6 shows peripheral T-cell receptor quantitation and clonality metrics at baseline and cycle 2 day 1 by mutation subgroup.
Data Availability Statement
Upon request, and subject to review, Pfizer will provide the data that support the findings of this study. Subject to certain criteria, conditions, and exceptions, Pfizer may also provide access to the related individual deidentified participant data. See https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information.