Abstract
Biomolecular condensates are dynamic structures formed through diverse mechanisms, including liquid-liquid phase separation. These condensates have emerged as crucial regulators of cellular processes in eukaryotic cells, enabling the compartmentalization of specific biological reactions while allowing for dynamic exchange of molecules with the surrounding environment. RNA silencing, a conserved gene regulatory mechanism mediated by small RNAs (sRNAs), plays pivotal roles in various biological processes. Multiple types of biomolecular condensate, including dicing bodies, processing bodies, small interfering RNA bodies, and Cajal bodies, have been identified as key players in RNA silencing pathways. These biomolecular condensates provide spatial compartmentation for the biogenesis, loading, action, and turnover of small RNAs. Moreover, they actively respond to stresses, such as viral infections, and modulate RNA silencing activities during stress responses. This review summarizes recent advances in understanding of dicing bodies and other biomolecular condensates involved in RNA silencing. We explore their formation, roles in RNA silencing, and contributions to antiviral resistance responses. This comprehensive overview provides insights into the functional significance of biomolecular condensates in RNA silencing and expands our understanding of their roles in gene expression and stress responses in plants.
Introduction
Cells are highly organized and compartmentalized to facilitate the numerous chemical reactions they host. In addition to membrane-bounded organelles, cells also possess membraneless compartments/organelles, also known as biomolecular condensates, that significantly contribute to cellular organization and function (Banani et al. 2017; Boeynaems et al. 2018; Zhao and Zhang 2020). Biomolecular condensates are dynamic and liquid-like in nature, harboring a concentrated mix of diverse biomolecules, including proteins, nucleic acids (DNAs and RNAs), and others. These structures are capable of undergoing fusion and separation (Brangwynne et al. 2009; Hyman et al. 2014; Gomes and Shorter 2019; Ismail et al. 2021). Differing from membrane-bound organelles, biomolecular condensates enable rapid exchange of components with their surroundings. Moreover, they possess the ability to assemble and disassemble in response to stimuli, thereby influencing cellular responses to environmental stresses (Alberti and Hyman 2021; Xu et al. 2021). Liquid-liquid phase separation (LLPS) has been considered as one of the critical driving forces behind biomolecular condensate formation. LLPS refers to the spontaneous separation of homogenous mixture into 2 or more distinct liquid phases with kinetic and thermodynamic alteration (Hyman et al. 2014; Brangwynne et al. 2015; Alberti and Dormann 2019). In certain instances, these liquid-like condensates can even transit into solid-like or gel-like structures over time (Banani et al. 2017; Shin and Brangwynne 2017). LLPS is facilitated by multivalent interactions among different components, often involving proteins with intrinsically disordered regions (IDRs), prion-like domains (PrLDs), and/or DNA/RNA binding domain, as well as RNA molecules (Alberti et al. 2019; Gomes and Shorter 2019; Martin and Holehouse 2020; Borcherds et al. 2021). Local concentrations of constituents, modification of protein/RNA components, and ATPase energy-dependent regulation collectively influence the LLPS capability and the homeostasis of biomolecular condensates (Gomes and Shorter 2019; Ismail et al. 2021). In addition, other forces also contribute to the assembly of biomolecular condensates. One such force is percolation, where biomolecules with high multivalency come together through processes like oligomerization or interactions between proteins or nucleic acids, thus forming condensates (Tibble and Gross 2023).
Biomolecular condensates play critical roles in various biological processes within eukaryotes (Banani et al. 2017; Emenecker et al. 2021; Xu et al. 2021). RNA molecules are not only major components of numerous biomolecular condensates but also act as essential regulators impacting their dynamics and functions (Rhine et al. 2020). These condensed structures, often termed RNA granules, play crucial roles across the entire spectrum of RNA's life cycle, including transcription, splicing, processing, modification, translation, and quality control of RNA molecules (Gomes and Shorter 2019; Lin and Fang 2021). In plants, biomolecular condensates have emerged as key players in regulating various aspects of growth and development, such as phytohormone- and photosynthesis-mediated growth and development (Wunder et al. 2018; Atkinson et al. 2020; He et al. 2020; Ouyang et al. 2020; Song et al. 2021; Cairo et al. 2022; Chen et al. 2022; Jing et al. 2022; Zhang et al. 2022; Kuan et al. 2023), water homeostasis (Dorone et al. 2021), osmotic stress sensing (Wang et al. 2022), response to heat or cold stress (Jung et al. 2020; Tong et al. 2022; Zhu et al. 2022), pathogen resistance (Huang et al. 2021; Wang and Gu 2022), and RNA silencing (Li et al. 2021b; Xie et al. 2021; Han et al. 2023). Notably, a significant proportion of these plant biomolecular condensates are closely associated with RNA, highlighting their crucial roles in RNA-related processes.
The RNA silencing pathway, a highly RNA-involved process, operates in coordination with various cellular condensates. In plants, a range of endogenous small RNA (sRNA) classes exists, including microRNAs (miRNAs), trans-acting small interfering RNAs (ta-siRNAs, also known as phased siRNAs or phasiRNAs), heterochromatic small interfering RNAs (hc-siRNAs), and natural antisense siRNAs (nat-siRNAs) (Huang et al. 2016; Song et al. 2019). These sRNAs are crucial components of the RNA silencing mechanism, and their biogenesis and action are closely linked to RNA-associated processes that potentially benefit from subcellular compartmentation. As an example, dicing bodies (D-bodies) serve as sites for miRNA biogenesis, pivotal in the processing of miRNAs by the dicing complex (Fang and Spector 2007; Xie et al. 2021). Processing bodies (P bodies), well characterized as locales for mRNA decay and translation inhibition (Maldonado-Bonilla 2014), have recently been observed to correlate with miRNA action and turnover (Solis-Miranda et al. 2023). Moreover, cytoplasmic siRNA bodies have been shown to integrate key components for siRNA biogenesis (Jouannet et al. 2012; Tan et al. 2023). As for hc-siRNA biogenesis, the presence of Dicer-like endonuclease 3 (DCL3) and RNA-dependent RNA polymerase 2 (RDR2) within Cajal body-like structures suggests potential connections between hc-siRNA production and these compartments (Li et al. 2006; Pontes et al. 2006).
In this review, we aim to provide a comprehensive overview of the pivotal role played by biomolecular condensates in RNA silencing pathways. We intend to summarize the current literature on the formation mechanisms and the functions of these biomolecular condensates in the biogenesis, loading, and action of various types of sRNAs. Additionally, we will explore the regulatory mechanisms that govern these biomolecular condensates in plant responses to different stress, with a special emphasis on viral infections. By synthesizing the existing research, we hope to shed light on the intricate interplay existing between biomolecular condensates and RNA silencing, offering insights into the significance of these condensates in plant defense against viral pathogens.
D-body and miRNA biogenesis
D-body serves as site for pri-miRNA processing
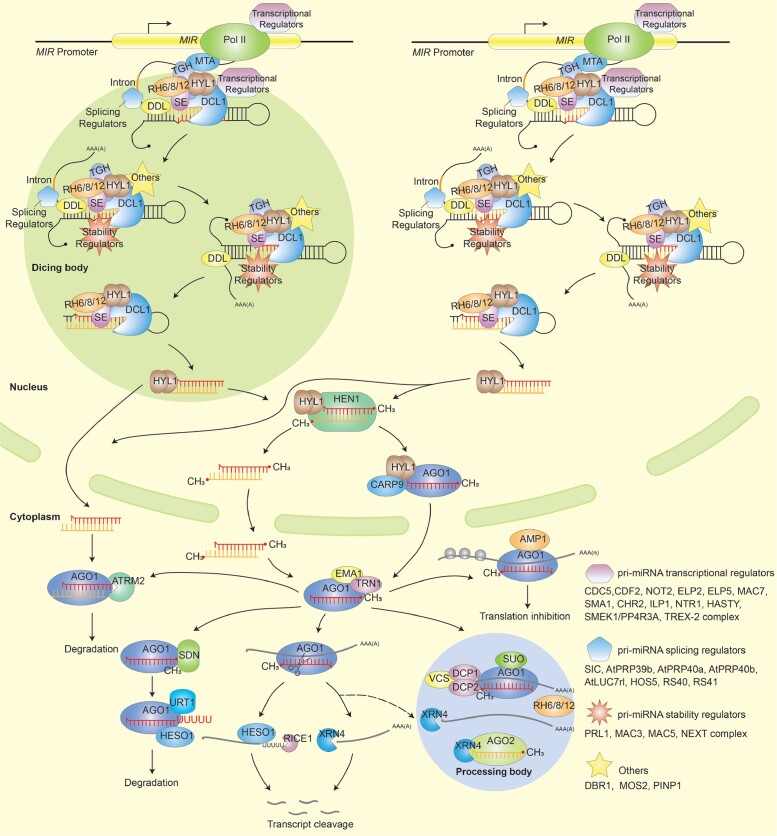
The process of miRNA biogenesis is initiated through the transcription of MIR genes by RNA polymerase II, resulting in the synthesis of primary miRNAs (pri-miRNAs) (Xie et al. 2005; Kim et al. 2011). The Dicing complex, which comprises DCL1, the double-stranded RNA-binding protein Hyponastic leaves 1 (HYL1), and the C2H2-type zinc finger protein SERRATE (SE), is responsible for processing pri-miRNAs into mature miRNAs (Fang and Spector 2007; Song et al. 2007).
In 2004, a pivotal discovery by Han et al. revealed that in Arabidopsis, GFP-fused HYL1 predominantly localizes within subnuclear small bodies as well as ring-like structures (Han et al. 2004). This study also prompted speculation about the potential occurrence of miRNA metabolism within the nucleus, possibly in association with perinucleolar RNA-processing bodies. Subsequent investigations conducted in 2007 further substantiated the subcellular localization of DCL1 and HYL1 within nuclear bodies distributed in nucleoplasm (Fang and Spector 2007). These nuclear bodies, termed D-bodies, exhibit a unique distribution pattern distinct from that of Cajal bodies. However, SE exhibited a heterogeneous subnuclear pattern. This study also suggested that these proteins interact in D-bodies. These observations underscore the potential roles of D-bodies in pri-miRNA processing. Around the same time, Song et al. identified the co-localization of DCL1 and HYL1 in a nuclear body distinct from the Cajal bodies (Song et al. 2007). Fujioka also discovered that DCL1, HYL1, and SE form nuclear bodies located close to, yet distinct from, nucleoli (Fujioka et al. 2007). Importantly, D-bodies have been shown to harbor pri-miR163 and pri-miR173, underscoring their roles as sites for pri-miRNA processing (Fang and Spector 2007; Fujioka et al. 2007).
Subsequently, numerous proteins that interact with DCL1, HYL1, and/or SE through genetic and biochemical approaches have been reported to be involved in D-body accumulation (Table 1). Dysfunction of proteins such as NOT2 (Wang et al. 2013) or PINP1 (Qiao et al. 2015) leads to an increased number of D-bodies, while mutations in MOS2 (Wu et al. 2013), ELP2 (Fang et al. 2015), ELP5 (Fang et al. 2015), MAC7 (Jia et al. 2017), SMEK1/PP4R3A (Su et al. 2017; Wang et al. 2019b), MAC3 (Li et al. 2018a), or TREX-2 complex (Zhang et al. 2020) result in a reduction in the number of D-bodies. However, the precise functions of these proteins in the formation of D-bodies remain to be fully understood.
Table 1.
List of proteins that function in miRNA biogenesis
| Proteins | Interacting protein | Function | Localization | Dicing body accumulation in mutants | Reference | |
|---|---|---|---|---|---|---|
| pri-miRNA transcriptional regulators | CDF2 | DCL1 and HYL1 | Regulates transcription and processing of pri-miRNAs | Dicing body | Unknown | (Sun et al. 2015) |
| CDC5 | DCL1 and SE | Recruits RNA Pol II to MIR promoters | Dicing body | Unknown | (Zhang et al. 2013) | |
| CHR2 | SE | Promotes transcription of pri-miRNAs and remodels secondary structures of pri- miRNAs | Dicing body | Unknown | (Wang et al. 2018b) | |
| ELP2 and ELP5 | DCL1 and SE | Recruits RNA Pol II to MIR promoters and DCL1 to MIR genes | Dicing body | Decreased | (Fang et al. 2015) | |
| HASTY | DCL1 | Promotes transcription and processing of pri-miRNAs | Dicing body | Unknown | (Cambiagno et al. 2021) | |
| ILP1 and NTR1 | DCL1 and SE | Regulates Pol II transcription elongation | Dicing body | Unchanged | (Wang et al. 2019a) | |
| MAC7 | HYL1 | May affect MIR transcription elongation and HYL1 localization in Dicing bodies | Not concentrated in Dicing body | Decreased | (Jia et al. 2017) | |
| NOT2 | DCL1 and SE | Interacts with Dicing complex and recruits RNA Pol II and DCL1 | Dicing body | Increased | (Wang et al. 2013) | |
| SMA1 | DCL1 and SE | Recruits RNA Pol II to MIR promoters, affects MIR transcription and intron splicing of pri-miRNAs | Dicing body | Unknown | (Li et al. 2018b) | |
| SMEK1/PP4R3A | HYL1 | Dephosphorylates HYL1 and recruits Pol II to MIR promoters | Dicing body | Decreased | (Su et al. 2017; Wang et al. 2019b) | |
| RH6, RH8, RH12 | DCL1, HYL1 and SE | Promotes dicing body formation and facilitates Pol II to bind MIR genes | Dicing body | Decreased | (Li et al. 2021b) | |
| TREX-2 Complex | SE | Recruits RNA Pol II to MIR promoters, regulates transcription, processing, and nuclear export | Nucleus | Decreased | (Zhang et al. 2020) | |
| pri-miRNA splicing regulators | SIC | Unknown | Regulates pri-miRNA processing and facilitates splicing of pri-miRNAs | Dicing body | Unknown | (Zhan et al. 2012) |
| HOS5, RS40 and RS41 | HYL1 and SE | Regulates miRNA strand selection and miRNA levels maintenance | Splicing speckles | Unknown | (Chen et al. 2015) | |
| AtPRP39b, AtPRP40a, AtPRP40b and AtLUC7rl | SE | Regulates pri-miRNA processing | Unknow | Unknown | (Knop et al. 2017) | |
| SEAP1 | SE | Promotes interaction of DCL1 complex with pri-miRNAs and affects pri-miRNA splicing | Nucleus | Unknown | (Li et al. 2021a) | |
| pri-miRNA stability regulators | DDL | DCL1 | Regulates pri-miRNA stability | Unknow | Unknown | (Yu et al. 2008) |
| PRL1 | DCL1, HYL1 and SE | Regulates pri-miRNA stability | Dicing body | Unknown | (Zhang et al. 2014) | |
| MAC3 | DCL1 and SE | Regulates pri-miRNA stability | Dicing body | Decreased | (Li et al. 2018a) | |
| MAC5 | SE | Regulates pri-miRNA stability | Dicing body | Unknown | (Li et al. 2020) | |
| NEXT Complex | SE | Regulates degradation of pri-miRNAs | Unknown | Unknown | (Bajczyk et al. 2020; Gao et al. 2020) | |
| Others | DBR1 | Unknown | Regulates pri-miRNA binding to DCL1 and HYL1 | Not localized in Dicing body | Increased | (Li et al. 2016) |
| MOS2 | pri-miRNAs | Regulates pri-miRNA processing | Not localized in Dicing body | Decreased | (Wu et al. 2013) | |
| MTA | TGH | Is required for co-localization of DCL1/HYL1 with Pol II | Unknown | Unknown | (Bhat et al. 2020) | |
| PINP1 | pri-miRNAs | Facilitates correct subnuclear localization of processing complex | In nucleus | Increased | (Qiao et al. 2015; Gui et al. 2022) | |
| TGH | DCL1, HYL1 and SE | Promotes DCL1 cleavage efficiency and/or recruitment of pri-miRNAs | Dicing body | Unknown | (Ren et al. 2012) | |
| SAID1, SAID2 | pri-miRNAs and SE | Hijacks pri-miRNA processing and promotes phosphorylation and destabilization of SE | In nucleus | Unknown | (Shang et al. 2023) | |
| CYP71 | SE | Promotes miRNA processing | Dicing body and nucleoplasm | Decreased | (Zhao et al. 2023) | |
Recent research has brought valuable insights into the formation and function of D-bodies in pri-miRNA processing. It has been discovered that D-bodies are formed through the phase separation of SE (Fig. 1) (Li et al. 2021b; Xie et al. 2021). SE possesses 3 IDRs, with the N-terminal IDR enabling SE to undergo phase separation both in vitro and in vivo. In contrast, neither DCL1 nor HYL1 exhibits the capacity to form droplets in vitro (Li et al. 2021b; Xie et al. 2021). Notably, mutation in SE or deletion of the N-terminal IDR hinders the formation of D-bodies both in vitro and in vivo (Xie et al. 2021). Importantly, a variant of SE lacking the N-terminal IDR exhibits reduced ability to facilitate miRNA processing within an in vitro reconstituted system. Furthermore, this mutation leads to decreased miRNA accumulation in transgenic plants. Conversely, the processing products, miRNA/miRNA* duplexes, along with HYL1, are exported from D-bodies in vitro. This observation supports the notion that D-bodies function as miRNA processing centers (Xie et al. 2021).
Figure 1.
The interplay of biomolecular condensates with miRNA expression and processing. Initially, pri-miRNAs are transcribed by RNA Pol II and subsequently processed into miRNAs. D-body formation relies on core components such as DCL1, HYL1, SE, and RNA helicases RH6/8/12. Notably, both SE and RH6/8/12 contain IDR regions. SE undergoes LLPS and incorporates DCL1, HYL1, and pri/pre-miRNAs, thereby facilitating the formation of D-bodies. RH6/8/12 interacts with SE and promotes its phase separation, facilitating D-body formation. Additionally, various other proteins involved in regulating pri-miRNA transcription, stability, and maturation interact with D-body components and may also localize in and influence D-body formation. Following processing, miRNA duplexes are methylated by HEN1 and then either sorted into AGO1 or transported out of the nucleus for cytoplasmic loading into AGO1. In the cytoplasm, miRNA deceases target gene expression by mRNA cleavage and/or translation inhibition. The translation inhibition processes of miRNAs take places on ER. The cleavage products are partially degraded, possibly by XRN4 within P bodies. Additionally, SUO and VARICOSE may facilitate the translation inhibition activity of miRNAs within P bodies. The turnover of unmethylated miRNAs is likely fulfilled by ATRM2 in P bodies and/or related biomolecular condensates. Mature miRNAs are predominantly degraded through SDN1-3, URT1, and HESO1-mediated degradation within P bodies and/or related biomolecular condensates. Furthermore, XRN4 participates in the degradation of miRNA* in P bodies.
Additionally, the study conducted by Li et al. has uncovered another significant aspect of D-bodies (Li et al. 2021b). They identified that RNA helicase 6 (RH6), RH8, and RH12, a group of DEAD-box RNA helicases family, interact with SE and promote the phase separation of SE in vitro, as well as the formation of D-bodies in vivo (Li et al. 2021b). Malfunction of RH6/8/12 results in decreased accumulation of D-bodies and reduced levels of pri-miRNAs and miRNAs (Li et al. 2021b). This observation highlights the essential involvement of these RNA helicases in the proper formation of D-bodies, contributing to the efficiency of miRNA biogenesis. Moreover, Cyclophilin71 (CYP71), a peptidyl-prolyl isomerase, interacts with SE and promotes its phase separation, which in turn facilitates the formation of D-bodies and enhances the activity of the Dicing complex (Zhao et al. 2023).
Moreover, the work of Xie et al. has presented evidence that highlights the advantageous role of biomolecular condensates in miRNA biogenesis (Xie et al. 2021). In vitro experiments involving the combination of SE with DCL1, HYL1, and pre-miRNAs have demonstrated that SE droplets can incorporate these client proteins, as well as pre-miRNAs into the formed droplets (Xie et al. 2021). The incorporation of various miRNA processing-related factors underscores the advantageous impact of D-bodies' condensation on miRNA processing. Indeed, the processing of pre-miRNAs exhibits a positive correlation with the formation of SE droplets in vitro. And in vivo, the decreased and rescued miRNA accumulation in transgenic lines with SE carrying deleted IDRs or substituted IDRs further highlight the functional role of phase-separated D-bodies in miRNA processing. These findings provide evidence for the functional significance of phase-separated D-bodies in facilitating effective miRNA processing. Furthermore, the incorporation of HYL1 into D-bodies and its subsequent release from these structures after processing imply the material exchange of D-bodies with their surroundings (Xie et al. 2021). This phenomenon aligns with the liquid-like characteristics of biomolecular condensates, which enable rapid substance exchange with the cellular environment.
pri-miRNA transcription couples with processing in D-bodies?
The current understanding of D-bodies suggests that their role extends beyond miRNA processing and may encompass additional steps in miRNA biogenesis. One crucial step of miRNA biogenesis is the transcription of MIR genes, and increasing studies propose that the transcription of pri-miRNA might be orchestrated with processing within D-bodies. Recent research has provided direct evidence supporting the co-transcriptional processing of pri-miRNAs (Fig. 1) (Gonzalo et al. 2022). For pri-miRNAs that undergo processing from the loop to the base, their entire biogenesis occurs co-transcriptionally (Gonzalo et al. 2022). For pri-miRNAs processed from the base to the loop, their biogenesis requires a subsequent posttranscriptional process within the nucleoplasm. The authors propose that the posttranscriptional processing of pri-miRNAs that processed from base to loop may occur within D-bodies.
A range of accessory factors has been identified to simultaneously regulate MIR transcription and miRNA processing (Table 1; Fig. 1). Most of these proteins localize and/or interact with Dicing complexes in D-bodies (Table 1). For example, the Elongator complex interacts with the Dicing complex and recruits RNA Pol II to MIR promoters and DCL1 to MIR genes, highlighting its role in coupling transcription and processing of pri-miRNAs (Fang et al. 2015). This study also found that components of the Elongator complex, such as ELP2 and ELP5, are necessary for both MIR transcription and pri-miRNA processing, suggesting that these 2 processes are coupled by elongator complex. The association of DCL1 with the chromatin of MIR loci also suggests a potential coupling between transcription and processing of pri-miRNAs (Fang et al. 2015). NOT2, a general factor that promotes transcription, associates with both Pol II and pri-miRNA processing complexes, bridging MIR transcription and pri-miRNA processing (Wang et al. 2013). HASTY interacts with both DCL1 and MED37 (a subunit of mediator complex required for pri-miRNA transcription), facilitating the recruitment of DCL1 to MIR loci and linking pri-miRNA transcription and processing (Cambiagno et al. 2021). SMEK1/PP4R3A, a chromatin associated protein, interacts with Pol II at MIR genes to promote MIR transcription (Wang et al. 2019b). Simultaneously, it interacts with and dephosphorylates HYL1, stabilizing HYL1 in D-bodies and ultimately promoting miRNA biogenesis (Su et al. 2017). CDF2 also associates with Dicing complex and affects both the transcription and processing of pri-miRNAs (Sun et al. 2015). Additionally, proteins such as CDC5 (Zhang et al. 2013), SMA1 (Li et al. 2018b), CHR2 (Wang et al. 2018b), TREX-2 complex (Zhang et al. 2020), and RH6/8/12 (Li et al. 2021b) interact with the Dicing complex and promote MIR transcription. Some of these proteins have been reported to regulate MIR transcription by influencing the binding of RNA polymerase II to MIR loci. Furthermore, 2 members of the spliceosome disassembly complex, ILP1 and NTR1, associate with the Dicing complex and promote MIR transcription by facilitating the transcription elongation of MIR genes (Wang et al. 2019a). MAC7 interacts with HYL1 and may promote the transcription elongation of MIR genes (Jia et al. 2017). Although MAC7 does not localize in D-bodies, it affects the localization of HYL1 in D-bodies, potentially linking MIR transcription and pri-miRNA processing. Therefore, D-bodies, which may house multiple transcriptional regulators, could potentially orchestrate MIR transcription and pri-miRNA processing.
Moreover, splicing factors have also been demonstrated to exhibit correlation with components of the dicing complex, simultaneously affecting pri-miRNA maturation. For example, SMA1, which interacts with Dicing complex and promotes MIR transcription, also facilitates the intron splicing of pri-miRNAs (Table 1) (Li et al. 2018b). SICKLE colocalizes with HYL1 in D-bodies and facilitates the splices of specific pri-miRNAs (Zhan et al. 2012). Some other proteins linked to RNA splicing, such as HOS5, RS40, RS41 (Chen et al. 2015), and U1 snRNP auxiliary proteins AtPRP39b, AtPRP40a, AtPRP40b, and AtLUC7rl (Knop et al. 2017; Stepien et al. 2022), have been found to colocalize and/or interact with Dicing complex and stimulate pri-miRNA processing. Another example is Serrate-Associated Protein 1 (SEAP1), which interacts with SE. SEAP1 facilitates interactions between dicing complex and pri-miRNAs, influencing the splicing of specific portions of pri-miRNAs (Li et al. 2021a). These findings collectively shed light on functional significance of D-bodies as specialized compartments dedicated to efficient pri-miRNA maturation.
Additional functions of D-body in miRNA biosynthesis
In addition to co-transcriptional processing and maturation of pri-miRNAs, D-bodies may regulate the stability and posttranscriptional modification of pri-miRNAs and other related procedures (Fig. 1). Several proteins related to RNA splicing, including MAC components such as PRL1 (Zhang et al. 2014), MAC3 (Li et al. 2018a), and MAC5 (Li et al. 2020), interact with Dicing complex within D-bodies and affect pri-miRNA stability. The nuclear exosome targeting complex, involved in RNA quality control, interacts with SE and triggers the degradation of pri-miRNAs (Bajczyk et al. 2020; Gao et al. 2020). DDL, a fork head-associated domain protein that is predicted to mediate protein-protein interactions, interacts with DCL1 and affects pri-miRNA stability (Yu et al. 2008). SAID1/2, which contain IDR, sequester pri-miRNAs from SE and aggregate with SE, promoting its phosphorylation and destabilization (Shang et al. 2023).
Additionally, TGH, which is predicted to be involved in RNA binding or processing, associates with Dicing complex within D-bodies and promotes DCL1 cleavage efficiency and/or the recruitment of pri-miRNAs (Ren et al. 2012). The mRNA adenosine methylase (MTA) is responsible for writing m6A on pri-miRNAs (Bhat et al. 2020). A study in mammalians showed that the m6A modification on mRNA is co-transcriptional (Slobodin et al. 2017). In plants, MTA interacts with both TGH and Pol II, and MTA is required for the co-localization of DCL1/HYL1 with Pol II (Bhat et al. 2020). As some of these proteins also localize and/or interact with Dicing complex in D-bodies (Table 1), these results indicate that m6A modification is likely to occur within D-bodies. Interestingly, it is suggested that D-bodies may function as storage sites for dicing complexes (Gonzalo et al. 2022).
Of note, a mutation of DBR1, the only known RNA lariat debranching enzyme, significantly increases the number of D-bodies. This mutation results in the accumulation of excess intron-derived lariat RNAs, which can compete with pri-miRNAs for binding to DCL1 and HYL1 (Table 1) (Li et al. 2016). Consistent with the association of lariat RNA with DCL1 and HYL1, lariat RNAs have been observed to partially co-localize with HYL1 in D-bodies within plant cells (Li et al. 2016).
After the processing of miRNAs, the formed miRNA/miRNA* duplexes can exit D-bodies with the assistance of HYL1 (Fig. 1) (Xie et al. 2021). Certain nuclear-cytoplasm transporting factors, such as TREX-2 and NUP1 (Zhang et al. 2020), have been shown to interact with the Dicing complex, indicating the storage of transporting factors within D-bodies.
Dynamics of D-bodies under stresses and their contribution to stress responses
As a biomolecular condensate that formed by LLPS, the assembly and disassembly of D-bodies are dynamically regulated to coordinate plant growth and stress responses. Accumulating evidence has revealed that the concentration of key D-body components undergoes dynamic changes in response to various stresses. These changes can affect the homeostasis of D-body components inside and subsequently influence the dynamics of D-bodies. For instance, studies have demonstrated that the concentration of RH6/8/12 can modulate the formation of D-bodies (Fig. 1) (Li et al. 2021b). The VPg protein of Turnip mosaic virus hijacks RH6/8/12, resulting in decreased accumulation of RH6/8/12 in the nucleus and ultimately a reduction in the number of D-bodies (Huang et al. 2010; Li et al. 2021b). The dynamic regulation of D-body may, in turn, contribute to plant-virus interaction. Research has shown that decreased accumulation of D-bodies is associated with attenuated plant antiviral resistance (Table 2) (Li et al. 2021b). RH6/8/12 proteins are then enriched near cytoplasmic viral replication sites, facilitating the formation of membraneless V-bodies by promoting phase separation of VPg protein of viruses (Li et al. 2021b). These V-bodies facilitate viral proliferation by acting as compartments that concentrate molecules required for proliferation and/or protecting viral components from the host immune systems. Similar compartments formed by other viruses such as Potato virus A (Hafren et al. 2015), Barley yellow striate mosaic virus (Fang et al. 2022), and Pea enation mosaic virus 2 (Brown et al. 2021) are also involved in regulating their proliferations and other biological processes (Table 3). In addition to biotic stresses, environmental stresses also impact the homeostasis of D-bodies. During the transition from dark to red light or from red light to dark, the stability of DCL1 protein is modulated by the interacting transcription factor phytochrome-interacting factor 4 (Sun et al. 2018). Under strontium stress, pri-miRNA accumulation increases, whereas the HYL1 protein level decreases (Pyo et al. 2020). It would be interesting to investigate whether D-body formation/turnover is affected under these stressed conditions. Additionally, the number of D-bodies decreases after cell wall removal (Delenko et al. 2022).
Table 2.
List of biomolecular condensates-regulated upon viral infections
| Biomolecular condensates | Viral proteins take effects | Targeted host proteins | Organism | Roles of viral protein | Organelle accumulation alternation | Reference |
|---|---|---|---|---|---|---|
| D-body | VPg protein of Turnip mosaic virus | RH6, RH8, RH12 | Arabidopsis thaliana | Promotes viral proliferation | Decreased | (Li et al. 2021b) |
| P body | BV1 protein of Cabbage leaf curl virus | AS2 and DCP2 | Arabidopsis thaliana | Activates DCP2 decapping activity and siRNA production and promotes viral sensitivity of plant | Unknown | (Ye et al. 2015) |
| CP and MP of Tobacco mosaic virus | Unknown | Nicotiana benthamiana | Increases RNA decay rates for transgene and decreased transgene PTGS amplification | Increased size of P bodies and siRNA bodies | (Conti et al. 2017) | |
| HC-Pro protein of Potato Virus A | P0, AGO1, UBP1, VCS, eIF4E | Nicotiana benthamiana | Promotes formation of PVA-induced granules | Unknown | (Hafren et al. 2015) | |
| Unknown (Brome mosaic virus) | LSm1-7 complexes | Saccharomyces cerevisiae | Promotes viral translation and replication | Unknown | (Galão et al. 2010) | |
| Unknown (Cauliflower mosaic virus) | DCP5, VCS, and LSM1 | Arabidopsis thaliana | Relocates some of P body components for translation maintenance | Unknown | (Hoffmann et al. 2022) | |
| Unknown (Tobacco rattle virus) | Unknown | Arabidopsis thaliana | Induces translational repression of viral transcripts during viral recovery | Increased number of P bodies in recovered plants | (Ma et al. 2015) | |
| siRNA body | βC1 protein of Tomato yellow leaf curl China virus | NbCaM | Nicotiana benthamiana | Upregulates NbCaM expression and inhibit RDR6 expression | Unknown | (Li et al. 2014) |
| βC1 of Tomato yellow leaf curl China virus | NbCaM, NbSGS3 | Nicotiana benthamiana | NbCaM guides NbSGS3 into autophagy pathway for degradation | Reduces the number of NbSGS3 granules when NbCaM overexpressed | (Li et al. 2014; Li et al. 2017) | |
| p2 protein of Rice stripe virus | OsSGS3 | Oryza Sativa | Reduces ta-siRNA functions | Unknown | (Du et al. 2011) | |
| TAV of Cauliflower mosaic virus | Unknown | Arabidopsis thaliana | Impairs production of secondary siRNAs rather than primary siRNAs | Unknown | (Shivaprasad et al. 2008) | |
| TGBp1 of Plantago asiatica mosaic virus | SGS3 and RDR6 | Arabidopsis thaliana | Reduces ta-siRNA accumulation and changes subcellular localization of SGS3 | Reduces the aggregation of SGS3 bodies when suppressor activity crippled | (Okano et al. 2014) | |
| VISP1 (a host peptide induced by Cucumber mosaic virus) | SGS3 and autophagy-related protein 8 | Arabidopsis thaliana | Reduces biogenesis of ta-siRNAs and amplification of viral siRNAs | Decreased when co-expressed | (Tong et al. 2021) | |
| VPg protein of Potato virus A | SGS3 | Solanum tuberosum and Arabidopsis thaliana | VPg–SGS3 interaction benefits viral infection | Unknown | (Rajamaki et al. 2014) | |
| Cajal body | 16 K protein of Tobacco rattle virus | Coilin | Nicotiana benthamiana | Activates salicylic acid–dependent plant resistance | Decreased when co-expressed | (Shaw et al. 2019) |
| ORF3 protein of Groundnut rosette virus | Fibrillarin | Nicotiana benthamiana | Promotes long-distance movement of virus | Increased fusion of Cajal body-like structures with the nucleolus | (Kim et al. 2007a; Kim et al. 2007b) | |
| VPg protein of Potato virus A | Fibrillarin | Nicotiana benthamiana | Facilitates virus accumulation | Unknown | (Rajamaki and Valkonen 2009) | |
| V2 protein of Tomato yellow leaf curl virus | AGO4 | Nicotiana benthamiana | Suppresses antiviral methylation of AGO4 | Unknown | (Wang et al. 2020) |
Table 3.
List of biomolecular condensates formed by viruses
| Granules formed | Viral protein components | Host protein components | Organism | Functions | References |
|---|---|---|---|---|---|
| Virus bodies | VPg protein of Plum pox virus and Turnip mosaic virus | RH6/8/12 | Arabidopsis thaliana | Promotes viral proliferations | (Huang et al. 2010) |
| PVA-induced granules | HC-Pro protein of Potato Virus A | P0, AGO1, UBP1, VCS, eIF4E | Nicotiana benthamiana | Regulates viral translation | (Hafren et al. 2015) |
| Viral ribonucleoprotein complexes in nucleolar | p26 protein of Pea enation mosaic virus 2 | Fibrillarin | Nicotiana benthamiana | Prerequisite sites for systemic virus movement | (Brown et al. 2021) |
| Spherical granules | Phosphoprotein of Barley yellow striate mosaic virus | UBC32 | Barley | Promotes viral replication | (Fang et al. 2022) |
Posttranslational modifications can also regulate the stability of D-body components. Phosphorylation of HYL1 by SnRK2 kinase increases its accumulation in response to abscisic acid treatment (Yan et al. 2017). Conversely, in response to abscisic acid treatment, SMEK1/PP4R3A, the regulatory subunit of PP4, counteracts the phosphorylation of HYL1 by MPK3 and stabilizes HYL1 (Su et al. 2017). However, the impact of these protein modifications on the accumulation of D-bodies remains unknown.
P body: site for action and turnover of miRNAs?
P bodies, along with other biomolecular condensate, play important roles in the action and turnover of miRNAs. After processing, miRNA/miRNA* duplexes generated within D-bodies are carried out of D-bodies by HYL1 and loaded into Argonaute 1 (AGO1) (Xie et al. 2021). This loading process is facilitated by the interaction between HYL1 and AGO1 with a nuclear-localized protein called CARP9, which contains IDRs (Fig. 1) (Tomassi et al. 2020).
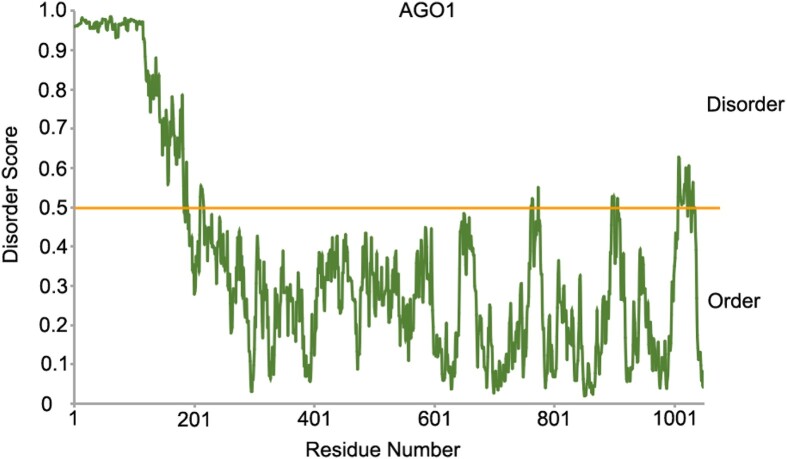
Biomolecular condensate has been found to be involved in the action of miRNAs and siRNAs. In human cells, AGO can form miRNA-induced silencing complex droplets with TNRC6B in the cytoplasm, which accelerates miRNA target deadenylation, leading to the downregulation of target gene expression (Sheu-Gruttadauria and MacRae 2018). Interestingly, both fly and human AGO can undergo lipid-mediated phase separation on the endoplasmic reticulum (ER) membrane and facilitates nascent-peptide ubiquitination, which thus couples posttranscriptional gene silencing with protein quality control (Gao et al. 2022). In plants, miRNA-loaded AGO1 inhibits the translation of miRNA targets on the ER (Li et al. 2013). The integral ER membrane protein AMP1 interacts with AGO1 and inhibits the association of mRNA target transcripts with membrane-bound polysomes (Fig. 1) (Li et al. 2013). Transiently expressed AGO1 has been observed to accumulate in cytoplasmic granules that colocalize with the ER (Li et al. 2013). It is worth noting that plant AGO1 also contains an IDR at the N terminal (Fig. 2). It would be interesting to determine whether plant AGO also undergo phase separation on the ER.
Figure 2.
Prediction of the disordered segments of plant AGO1 by IUPred2A. Protein residues with score above 0.5 is identified as disordered. The prediction was performed at web site version of IUPred2A using default parameters. The address is http://iupred2a.elte.hu.
P bodies are conserved biomolecular condensates that play important roles in RNA decay, RNA storage, and translation repression (Jang et al. 2020; Kearly et al. 2022). They are believed to be involved in sRNA-mediated gene silencing. The formation of P bodies is facilitated by proteins such as RH6/8/12 (Chantarachot et al. 2020). Regarding miRNA-mediated gene silencing, the exonuclease XRN4 has been implicated in the degradation of both 5′ and 3′ cleavage products generated by AGO1 slicing of target mRNAs (Fig. 1) (Souret et al. 2004; Ren et al. 2014). XRN4 interacts with AGO2 and forms cytoplasmic foci that co-localize with P bodies (Weber et al. 2008; Liu et al. 2020). Additionally, P body–localized proteins, such as VARICOSE (Brodersen et al. 2008), a component of the decapping complex, and SUO (Yang et al. 2012), a GW-repeat protein, have been identified as regulators of miRNA-mediated translational inhibition in Arabidopsis. However, it remains to be determined whether plant AGOs carry out target mRNA cleavage and translational inhibition within P bodies. Considering that plant AGO1 inhibits the translation of miRNA targets on the ER (Li et al. 2013) and human AGO1 undergoes phase separation on the ER (Gao et al. 2022), investigating the relationship between AGO granules on the ER and P bodies stands as an intriguing avenue for further exploration. In humans, the Importin β protein Imp8 has been shown to form a complex with AGO within P bodies, facilitating the delivery of AGO complexes to target mRNAs (Weinmann et al. 2009). In Arabidopsis, 2 importin β family proteins, EMA1 (Wang et al. 2011) and TRN1 (Cui et al. 2016), also have been implicated in the regulation of miRNA loading into AGO1 (Fig. 1). Moreover, TRN1 but not EMA1 has been shown to interact with AGO1 in both the nucleus and cytoplasm (Cui et al. 2016).
The turnover of miRNAs plays a crucial role in their homeostasis, and the integration of proteins and RNAs may involve biomolecular condensates. In Arabidopsis, unmethylated miRNA duplexes can be degraded by the exoribonuclease ATRIMMER 2 (ATRM2) in a 3′ to 5′ direction (Fig. 1) (Wang et al. 2018a). ATRM2 colocalizes and interacts with AGO1 in stress granules, biomolecular condensates often associated with P bodies, upon heat stress. For methylated miRNAs, proteins SDN1-3 are responsible for their truncation, both when free and bound to AGO1 (Ramachandran and Chen 2008; Yu et al. 2017; Chen et al. 2018). Following truncation, HESO1 and URT1 cooperate to uridylate the substrates of SDN1-3, thereby promoting their degradation (Fig. 1) (Ren et al. 2012; Zhao et al. 2012; Tu et al. 2015; Wang et al. 2015). HESO1 and URT1 colocalize and interact with AGO1 in cytoplasmic speckles in Nicotiana benthamiana (Ren et al. 2014; Wang et al. 2015). Considering that AGO1 is also found in P bodies and stress granules (Wang et al. 2018a; Solis-Miranda et al. 2023), these cytoplasmic speckles may represent P bodies and/or stress granules. Furthermore, AtXRN4, an exonuclease, contributes to the turnover of some miRNA*s, the passenger strand of the miRNA duplexes. Transiently expressed AtXRN4 interacts with AGO2 and forms granules within P bodies in N. benthamiana (Fig. 1) (Liu et al. 2020). A recent review has comprehensively summarized the presence of the previously mentioned components within P bodies. These components include various AGOs (such as AGO1, AGO2, AGO5, and AGO9), XRN4, and RH6/8/12 (Solis-Miranda et al. 2023). This compilation of evidence suggests a potential connection between P bodies and the regulation of miRNA turnover.
P bodies play significant roles in responses to abiotic and biotic stresses (Jang et al. 2019). Viruses, on the other hand, have developed various mechanisms to target specific components of P bodies and modulate their accumulation, thereby influencing their functions (Table 3) (Makinen et al. 2017). For example, infection of Tobacco mosaic virus in N. benthamiana leads to transcriptional up-regulation of genes associated with RNA decay, as well as an increase in the size of P bodies and siRNA bodies (Conti et al. 2017). Similarly, plants recovered from Tobacco rattle virus infection also display an increased number of P bodies (Ma et al. 2015). Although the exact impact of viruses on P body accumulation in other cases remains ambiguous, it is evident that the functions of P body are affected. For example, the HC-Pro protein of Potato Virus A has been found to relocate components of P bodies and stress granules to form of Potato Virus A-induced granules. The number of Potato Virus A-induced granules decreases when HC-Pro is impaired (Hafren et al. 2015). The BV1 protein of Cabbage leaf curl Virus activates the decapping activity of DCP2 and siRNA production and promotes plant susceptibility to viral infection (Ye et al. 2015). The VPg protein of Plum pox virus and Turnip Mosaic Virus interacts with RH6/8/12, core components that drive the formation of P bodies and D-bodies, and promotes virus proliferation (Huang et al. 2010; Chantarachot et al. 2020; Li et al. 2021b). Cauliflower mosaic virus relocates P body components DCP5, VCS, and LSM1 into viral factories, facilitating the translation of viral RNA (Hoffmann et al. 2022). Additionally, the decapping activator LSm1-7 complexes directly bind to the genomic RNA of Brome mosaic virus within P bodies, exerting regulatory effects that activate viral translation or promote viral replication (Galão et al. 2010).
siRNA body and ta-siRNA biogenesis
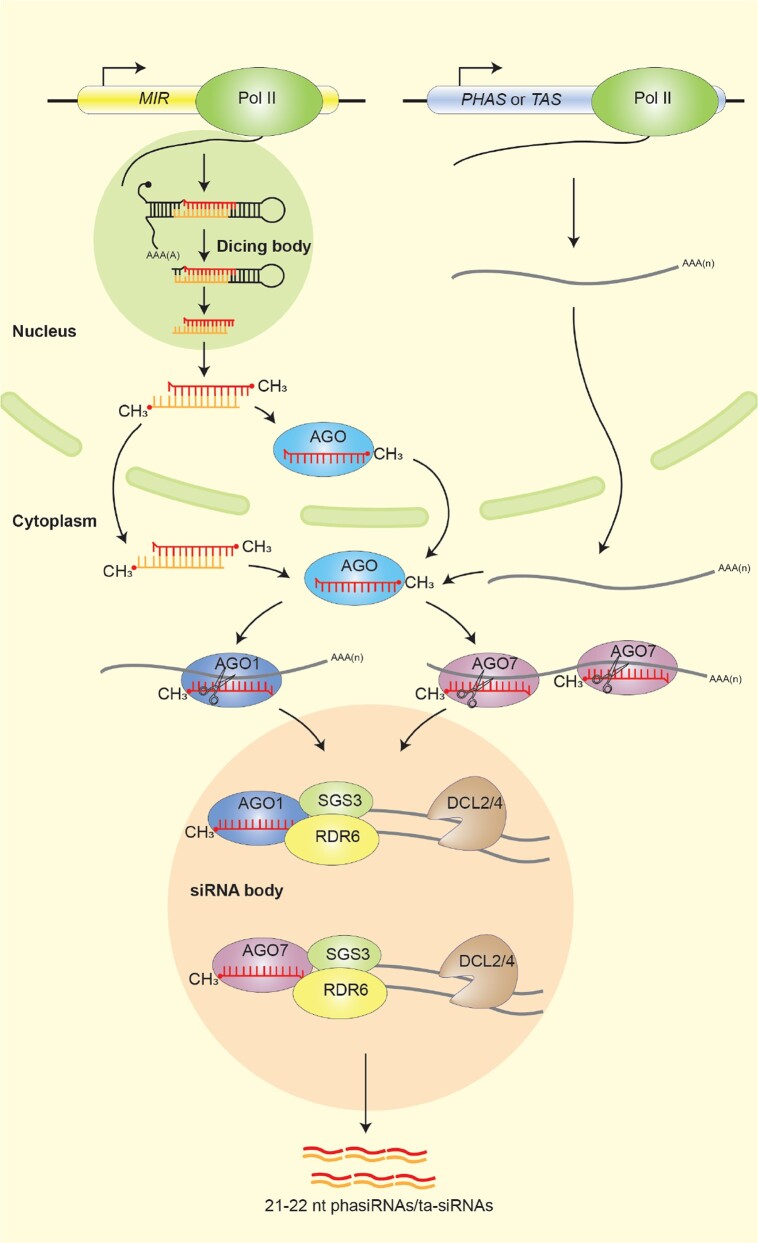
AGO7, SGS3, RDR6, and DCL4, the key components involved in ta-siRNA biogenesis, have been found to localize within a plant-specific cytoplasmic biomolecular condensate called the siRNA body, suggesting that this biomolecular condensate serves as a center for ta-siRNA biogenesis (Fig. 3) (Kumakura et al. 2009; Jouannet et al. 2012; Kim et al. 2021; Han et al. 2023; Tan et al. 2023). Unlike P bodies, these granules are primarily referred to as SGS3/RDR6 bodies due to the interaction and co-localization of SGS3 and RDR6 within them (Kumakura et al. 2009). Notably, SGS3, which contains a PrLD domain, is believed to play a critical role in the formation of siRNA bodies by LLPS (Kim et al. 2021). While full-length SGS3 can form cytoplasmic foci, SGS3 lacking the PrLD domain is evenly distributed in the nucleus (Kim et al. 2021). Further investigations have demonstrated that SGS3 interacts with RDR6 and drives the formation of siRNA bodies, which is necessary and sufficient for siRNA actions (Han et al. 2023; Tan et al. 2023). Moreover, the phosphorylation statue of SGS3 regulates the phase separation of SGS3 and the formation of SGS3/RDR6 bodies (Han et al. 2023). Furthermore, the phase separation of SGS3 contributes to plant antiviral immunity, and the phosphorylation status of SGS3 affect this immune response (Han et al. 2023). Both SGS3 and RDR6 possess RNA binding activity (Elkashef and Ding 2009; Kunej et al. 2021). Notably, RDR6 prefers to bind aberrant mRNAs without poly(A+) tails (Baeg et al. 2017), suggesting that RNA molecules may also regulate siRNA body formation. Indeed, RNAs targeted by SGS3 have been found to promote the formation of siRNA bodies (Tan et al. 2023).
Figure 3.
siRNA body and ta-siRNA biogenesis. phasiRNA-generating (PHAS) loci and ta-siRNA-generating (TAS) loci are transcribed by RNA Pol II. After miRNA-induced silencing complex biosynthesis, miRNAs that direct ta-siRNA biosynthesis are sorted in AGOs. They can fulfill transcript cleavage either in a “one-hit” manner with the AGO1 RISC complex or in a “two-hit” manner with the AGO7 RISC complex. The PrLD domain-containing protein SGS3, which interacts with RDR6, plays a crucial role in driving the formation of siRNA bodies through phase separation. Within siRNA bodies, the cleavage products of AGO1 and AGO7 are stabilized by SGS3 and converted into dsRNAs by RDR6. These dsRNAs are subsequently by DCL2/4 to produce ta-siRNAs and other phased-secondary siRNAs.
siRNA bodies play a pivotal role in plant resistance to viruses and are sensitive to viral infections and various stresses. The formation and dynamics of siRNA body can be influenced by multiple viruses (Table 2) (Makinen et al. 2017). For example, the p2 protein of Rice stripe virus interacts with rice SGS3 and reduce the functions of ta-siRNAs (Du et al. 2011). The βC1 protein of Tomato yellow leaf curl China virus upregulates the expression of calmodulin-like protein (NbCaM), an endogenous RNA silencing suppressor, and repress RDR6 expression (Li et al. 2014). Further studies have shown that NbCaM also negatively regulates siRNA body formation by interacting with SGS3 and guiding SGS3 into the autophagy pathway for degradation, thereby altering plant immunity (Li et al. 2017). The TGBp1 protein of Plantago asiatica mosaic virus interacts with SGS3 and RDR6 (Okano et al. 2014), altering the subcellular localization of SGS3 and the synthesis of dsRNA in the ta-siRNA pathway. Moreover, the virus-induced small peptide VISP1 interacts with both SGS3 and autophagy-related protein 8, promoting siRNA body degradation through the autophagy pathway and facilitating viral infection (Tong et al. 2021). The TAV protein of Cauliflower mosaic virus impairs the production of secondary but not primary siRNAs (Shivaprasad et al. 2008). In contrast, although the VPg protein of Potato virus A also interacts with SGS3 within siRNA body, the silencing of SGS3 actually reduces the accumulation of viral RNA (Rajamaki et al. 2014).
Interestingly, proteins associated with RNA quality control, which are predominantly concentrated within P bodies, also play a role in suppressing posttranscriptional gene silencing (Christie et al. 2011; Liu and Chen 2016; Kim 2023). For instance, factors related to both 5′-3′ and 3′-5′ cytoplasmic RNA decay pathways also repress endogenous and transgene posttranscriptional gene silencing (Zhang et al. 2015). Moreover, the key decapping enzyme DCP2 prevents transgene silencing in Arabidopsis (Thran et al. 2012). Notably, mutations in RNA Quality Control factors such as xrn4, dcp2, and vcs activate RDR6-dependent siRNAs targeting endogenous transcripts (Martinez de Alba et al. 2015). This observation raises an intriguing perspective on the interplay between the RNA silencing and RNA decay pathways, suggesting that when the quality control pathway is overwhelmed or compromised, decay substrates may spill into the RDR6/DCL2/DCL4 pathway, leading to sRNA biogenesis and subsequent RNA silencing processes (Christie et al. 2011; Liu and Chen 2016; Kim 2023).
Cajal body and heterochromatin siRNA production
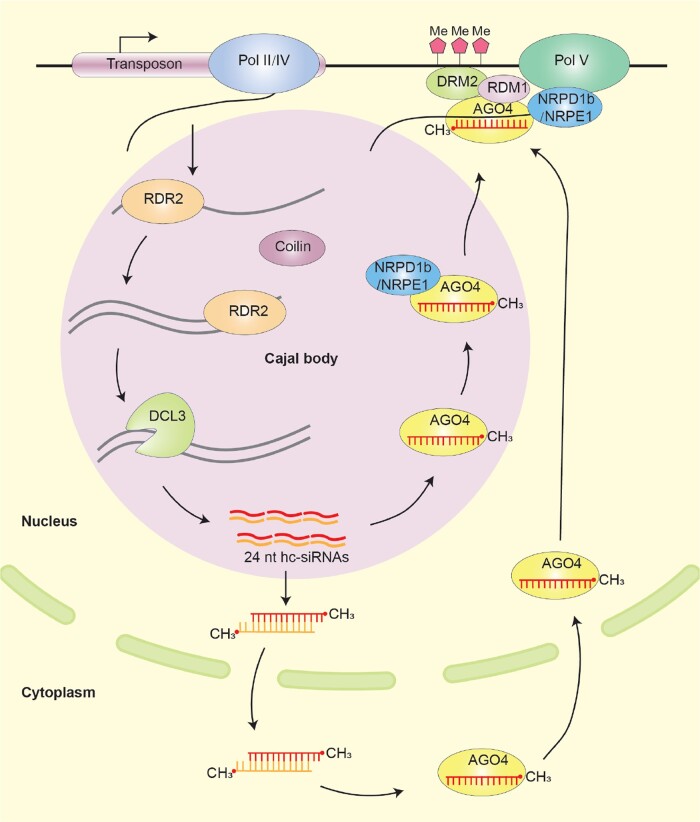
Cajal bodies are a type of nuclear biomolecular condensate involved in the processing and maturation of nuclear small non-coding RNAs, such as small nuclear RNA and small nucleolar RNA in yeast, animals, and plants (Machyna et al. 2013; Taliansky et al. 2023). These specialized organelles play regulatory roles in various biological processes, including transcription, splicing, ribosome biogenesis/rRNA maturation, and telomere maintenance (Love et al. 2017). In humans, more than 50 proteins have been identified to localize in Cajal bodies, while in plants, approximately 13 proteins have been observed to localize in Cajal bodies (Machyna et al. 2013; Love et al. 2017). While the involvement of Cajal bodies in hc-siRNA production in plants remains a subject of debate, several studies suggest their association with this process. RDR2, DCL3, AGO4, NRPD1b/NRPE1 of Pol IV/V, and 24-nt hc-siRNAs tend to be enriched in Cajal bodies (Li et al. 2006) or nucleolar dots that are likely to be Cajal bodies or related entities (Pontes et al. 2006) (Fig. 4). However, it should be noted that the DNA methyltransferase DRM2, which is essential for hc-siRNA function, is not localized in Cajal bodies. This suggests that Cajal bodies likely serve as the sites for hc-siRNA processing and storage rather than action (Li et al. 2006).
Figure 4.
Cajal body and heterochromatin siRNA production. The generation of 24-nt heterochromatin siRNA initiates with Pol IV/Pol II transcripts generated from transposable elements and repetitive DNA elements. These transcripts are converted into dsRNAs by RDR2 within Cajal bodies. Subsequently, DCL3 cleaves the dsRNAs, leading to the production of 24-nt hc-siRNAs. The hc-siRNAs are loaded into AGO4 complex within Cajal bodies and then transported to target sites for DNA methylation. Alternatively, the hc-siRNAs can also be exported into the cytoplasm for AGO4 assembly, and transported back into the nucleus to perform their functions. By base pairing with the ssRNA transcribed by Pol V, the AGO4-complex is recruited to the methylation site. It interacts with DRM2, RDM1, and NRPD1b/NRPE1, a Pol IV/Pol V subunit containing the PrLD domain. This complex confers RNA-dependent DNA methylation.
Coilin, an evolutionarily conserved protein, contains a typical IDR domain and plays a crucial role in the formation of Cajal bodies in both animals and plants (Collier et al. 2006; Navascues et al. 2008; Liu et al. 2009; Makarov et al. 2013; Courchaine et al. 2022). Additionally, NRPD1b/NRPE1, subunit protein of RNA polymerase IV and V that is involved in hc-siRNA production, also possesses a PrLD domain (Lei et al. 2021). Interestingly, AGO4 has been observed to localize in nuclear compartments other than Cajal bodies, suggesting the involvement of additional biomolecular condensates in hc-siRNA action (Li et al. 2008).
Cajal bodies, like other biomolecular condensates, display dynamic behavior influenced by various factors such as cell type, developmental stage, and life cycle in animals (Platani et al. 2000; Walker et al. 2009). In plants, Cajal bodies have been found to participate in stress response, particularly in plant-virus interactions (Table 2). Numerous studies have demonstrated interactions between viral proteins and components of Cajal bodies, shedding light on the viral manipulation of these organelles (Kim et al. 2007a; Rajamaki and Valkonen 2009; Semashko et al. 2012; Ruiz-Ruiz et al. 2013; Zheng et al. 2018; Shaw et al. 2019; Wang et al. 2020). For example, the 16-K protein of Tobacco rattle virus (TRV) interacts with Coilin, resulting in the delocalization of Coilin from Cajal body and the activation of salicylic acid-mediated resistance (Shaw et al. 2019). Similarly, the ORF3 protein of Groundnut rosette virus associates with fibrillarin (Kim et al. 2007a), another Cajal body component, leading to the reorganization of Cajal bodies and facilitating the systemic movement of the virus (Kim et al. 2007b). The VPg protein of Potato virus A interacts with fibrillarin in both nucleolar and Cajal bodies, promoting virus accumulation (Rajamaki and Valkonen 2009). Additionally, the V2 protein of Tomato yellow leaf curl virus interacts with AGO4 within Cajal bodies, suppressing its antiviral methylation activity (Wang et al. 2020). Furthermore, apart from virus-induced stress, Cajal bodies also participate in response to various abiotic stresses, such as hypoxia, salt stress, osmotic stress, UV irradiation, and heat shock. These roles were extensively discussed in a recently published review by Taliansky et al. (2023), providing comprehensive insights into the multifunctional roles of Cajal bodies in cellular biology.
Conclusion and future perspective
In conclusion, D-bodies, P bodies, siRNA bodies, and Cajal bodies have emerged as essential biomolecular condensates with demonstrated or predicted roles in regulating RNA silencing processes. The functional significance of biomolecular condensates has been extensively discussed in mammalian cells (Banani et al. 2017; Shin and Brangwynne 2017), providing valuable insights into their roles in RNA silencing and various other plant biological processes. These condensates can be categorized into 3 distinct functional roles: reaction crucible, sequestration, and organization hub (Shin and Brangwynne 2017). Biomolecular condensates can enhance cellular reactions by concentrating specific molecules within the condensates, thereby increasing reaction efficiency. This concept is illustrated by the enhancement of miRNA processing efficiency within D-bodies (Xie et al. 2021). Furthermore, biomolecular condensates may function as sequestration factors, aggregating specific components for future utilization. P bodies and stress granules, cytoplasmic condensates linked to translation complex storage, provide examples of this role, particularly during stress responses. These functions have been extensively reviewed by Solis-Miranda and colleagues (Solis-Miranda et al. 2023). Moreover, biomolecular condensates can contribute to spatial organization within cells. Cajal bodies, for example, are specifically associated with various RNA-related cellular processes, including small nuclear RNA and small nucleolar RNA biogenesis (Taliansky et al. 2023). However, whether and how the condensations truly facilitate RNA silencing processes remains to be fully elucidated.
The dynamic nature of biomolecular condensates enables them to rapidly respond to cellular stresses. Their ability to form and dissolve, along with their capacity to concentrate and organize specific components, contribute to the precise modulation of sRNA production and activities. These mechanisms enable cells to finely tune their stress responses and ensure the appropriate regulation of gene expression in diverse biological contexts. Future research is anticipated to unravel the intricate regulatory mechanisms that govern the formation and dynamic behavior of RNA silencing-related biomolecular condensates, and their impact on RNA silencing and stress responses. Investigations will aim to comprehend the mechanisms underlying their formation, dynamic regulation, factors influencing assembly and disassembly, as well as the interplay between different biomolecular condensates and RNA silencing pathways.
Acknowledgments
Our apology to our colleagues whose work could not be cited due to space restrictions. We thank Dr. Yijun Qi for his valuable contributions and constructive comments on the conceptualization and draft revisions of this manuscript.
Contributor Information
Qi Li, State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing, China.
Yang Liu, State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing, China.
Xiaoming Zhang, State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing, China; HainanYazhou Bay Seed Lab, Sanya, China.
Author contributions
X.Z. conceptualized the review outline. Q.L. and Y. L. curated information, prepared the figures, and wrote the original draft. X.Z. reviewed and edited the manuscript.
Funding
This work is supported by the National Key R&D Program of China (2022YFD1400800), National Natural Science Foundation of China (NSFC 32090010 and 32325042), Program of CAS (ZDBS-LY-SM027), and Hainan Yazhou Bay Seed Lab (B21HJ0104).
Data availability
The authors of distribution of materials integral to findings in this article according to the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Xiangming Zhang (zhangxm@ioz.ac.cn), Qi Li (liqi@ioz.ac.cn) and Yang Liu (liuyang2016@ioz.ac.cn). All data are incorporated into the article and its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Alberti S, Dormann D. Liquid-liquid phase separation in disease. Annu Rev Genet. 2019:53(1):171–194. 10.1146/annurev-genet-112618-043527 [DOI] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019:176(3):419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021:22(3):196–213. 10.1038/s41580-020-00326-6 [DOI] [PubMed] [Google Scholar]
- Atkinson N, Mao Y, Chan KX, McCormick AJ. Condensation of rubisco into a proto-pyrenoid in higher plant chloroplasts. Nat Commun. 2020:11(1):6303. 10.1038/s41467-020-20132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeg K, Iwakawa HO, Tomari Y. The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat Plants. 2017:3(4):17036. 10.1038/nplants.2017.36 [DOI] [PubMed] [Google Scholar]
- Bajczyk M, Lange H, Bielewicz D, Szewc L, Bhat SS, Dolata J, Kuhn L, Szweykowska-Kulinska Z, Gagliardi D, Jarmolowski A. SERRATE Interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic Acids Res. 2020:48(12):6839–6854. 10.1093/nar/gkaa373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017:18(5):285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SS, Bielewicz D, Gulanicz T, Bodi Z, Yu X, Anderson SJ, Szewc L, Bajczyk M, Dolata J, Grzelak N, et al. mRNA adenosine methylase (MTA) deposits m(6)A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2020:117(35):21785–21795. 10.1073/pnas.2003733117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018:28(6):420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021:67:41–50. 10.1016/j.sbi.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009:324(5935):1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nat Phys. 2015:11(11):899–904. 10.1038/nphys3532 [DOI] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008:320(5880):1185–1190. 10.1126/science.1159151 [DOI] [PubMed] [Google Scholar]
- Brown SL, Garrison DJ, May JP. Phase separation of a plant virus movement protein and cellular factors support virus-host interactions. PLoS Pathog. 2021:17(9):e1009622. 10.1371/journal.ppat.1009622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo A, Vargova A, Shukla N, Capitao C, Mikulkova P, Valuchova S, Pecinkova J, Bulankova P, Riha K. Meiotic exit in Arabidopsis is driven by P-body-mediated inhibition of translation. Science. 2022:377(6606):629–634. 10.1126/science.abo0904 [DOI] [PubMed] [Google Scholar]
- Cambiagno DA, Giudicatti AJ, Arce AL, Gagliardi D, Li L, Yuan W, Lundberg DS, Weigel D, Manavella PA. HASTY Modulates miRNA biogenesis by linking pri-miRNA transcription and processing. Mol Plant. 2021:14(3):426–439. 10.1016/j.molp.2020.12.019 [DOI] [PubMed] [Google Scholar]
- Chantarachot T, Sorenson RS, Hummel M, Ke H, Kettenburg AT, Chen D, Aiyetiwa K, Dehesh K, Eulgem T, Sieburth LE, et al. DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat Plants. 2020:6(6):675–685. 10.1038/s41477-020-0681-8 [DOI] [PubMed] [Google Scholar]
- Chen D, Lyu M, Kou X, Li J, Yang Z, Gao L, Li Y, Fan LM, Shi H, Zhong S. Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol Cell. 2022:82(16):3015–3029.e6. 10.1016/j.molcel.2022.05.026 [DOI] [PubMed] [Google Scholar]
- Chen J, Liu L, You C, Gu J, Ruan W, Zhang L, Gan J, Cao C, Huang Y, Chen X, et al. Structural and biochemical insights into small RNA 3′ end trimming by Arabidopsis SDN1. Nat Commun. 2018:9(1):3585. 10.1038/s41467-018-05942-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Cui P, Xiong L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015:43(17):8283–8298. 10.1093/nar/gkv751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M, Brosnan CA, Rothnagel JA, Carroll BJ. RNA Decay and RNA silencing in plants: competition or collaboration? Front Plant Sci. 2011:2:99. 10.3389/fpls.2011.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell. 2006:17(7):2942–2951. 10.1091/mbc.e05-12-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti G, Zavallo D, Venturuzzi AL, Rodriguez MC, Crespi M, Asurmendi S. TMV Induces RNA decay pathways to modulate gene silencing and disease symptoms. Plant J. 2017:89(1):73–84. 10.1111/tpj.13323 [DOI] [PubMed] [Google Scholar]
- Courchaine E, Gelles-Watnick S, Machyna M, Straube K, Sauyet S, Enright J, Neugebauer KM. The coilin N-terminus mediates multivalent interactions between coilin and Nopp140 to form and maintain Cajal bodies. Nat Commun. 2022:13(1):6005. 10.1038/s41467-022-33434-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Fang X, Qi Y. TRANSPORTIN1 Promotes the association of microRNA with ARGONAUTE1 in Arabidopsis. Plant Cell. 2016:28(10):2576–2585. 10.1105/tpc.16.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delenko K, Nuc P, Kubiak D, Bielewicz D, Dolata J, Niedojadlo K, Gorka S, Jarmolowski A, Szweykowska-Kulinska Z, Niedojadlo J. MicroRNA biogenesis and activity in plant cell dedifferentiation stimulated by cell wall removal. BMC Plant Biol. 2022:22(1):9. 10.1186/s12870-021-03323-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorone Y, Boeynaems S, Flores E, Jin B, Hateley S, Bossi F, Lazarus E, Pennington JG, Michiels E, De Decker M, et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell. 2021:184(16):4284–4298.e27. 10.1016/j.cell.2021.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Xiao D, Wu J, Jia D, Yuan Z, Liu Y, Hu L, Han Z, Wei T, Lin Q, et al. P2 of Rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol Plant Pathol. 2011:12(8):808–814. 10.1111/j.1364-3703.2011.00716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkashef S, Ding SW. Possible new RNA intermediate in RNA silencing. Nat Chem Biol. 2009:5(5):278–279. 10.1038/nchembio0509-278 [DOI] [PubMed] [Google Scholar]
- Emenecker RJ, Holehouse AS, Strader LC. Biological phase separation and biomolecular condensates in plants. Annu Rev Plant Biol. 2021:72(1):17–46. 10.1146/annurev-arplant-081720-015238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Cui Y, Li Y, Qi Y. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat Plants. 2015:1(6):15075. 10.1038/nplants.2015.75 [DOI] [PubMed] [Google Scholar]
- Fang XD, Gao Q, Zang Y, Qiao JH, Gao DM, Xu WY, Wang Y, Li D, Wang XB. Host casein kinase 1-mediated phosphorylation modulates phase separation of a rhabdovirus phosphoprotein and virus infection. Elife. 2022:11:e74884. 10.7554/eLife.74884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007:17(9):818–823. 10.1016/j.cub.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007:48(9):1243–1253. 10.1093/pcp/pcm099 [DOI] [PubMed] [Google Scholar]
- Galão RP, Chari A, Alves-Rodrigues I, Lobão D, Mas A, Kambach C, Fischer U, Díez J. LSm1-7 complexes bind to specific sites in viral RNA genomes and regulate their translation and replication. RNA. 2010:16(4):817–827. 10.1261/rna.1712910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Wang J, Jiang N, Zhang S, Wang Y, Zhang J, Li N, Fang Y, Yang L, Chen S, et al. Hyponastic leaves 1 protects pri-miRNAs from nuclear exosome attack. Proc Natl Acad Sci U S A. 2020:117(29):17429–17437. 10.1073/pnas.2007203117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhu Y, Wang H, Cheng Y, Zhao D, Sun Q, Chen D. Lipid-mediated phase separation of AGO proteins on the ER controls nascent-peptide ubiquitination. Mol Cell. 2022:82(7):1313–1328.e8. 10.1016/j.molcel.2022.02.035 [DOI] [PubMed] [Google Scholar]
- Gomes E, Shorter J. The molecular language of membraneless organelles. J Biol Chem. 2019:294(18):7115–7127. 10.1074/jbc.TM118.001192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo L, Tossolini I, Gulanicz T, Cambiagno DA, Kasprowicz-Maluski A, Smolinski DJ, Mammarella MF, Ariel FD, Marquardt S, Szweykowska-Kulinska Z, et al. R-loops at microRNA encoding loci promote co-transcriptional processing of pri-miRNAs in plants. Nat Plants. 2022:8(4):402–418. 10.1038/s41477-022-01125-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Zhang P, Wang D, Ding Z, Wu X, Shi J, Shen Q-H, Xu Y-Z, Ma W, Qiao Y. Phytophthora effector PSR1 hijacks the host pre-mRNA splicing machinery to modulate small RNA biogenesis and plant immunity. Plant Cell. 2022:34(9):3443–3459. 10.1093/plcell/koac176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren A, Lohmus A, Makinen K. Formation of Potato virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog. 2015:11(12):e1005314. 10.1371/journal.ppat.1005314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci U S A. 2004:101(4):1093–1098. 10.1073/pnas.0307969100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang X, Du R, Shan X, Xie D. The phase separation of SGS3 regulates antiviral immunity and fertility in Arabidopsis. Sci China Life Sci. 2023:66(8):1938–1941. 10.1007/s11427-022-2287-x [DOI] [PubMed] [Google Scholar]
- He S, Chou HT, Matthies D, Wunder T, Meyer MT, Atkinson N, Martinez-Sanchez A, Jeffrey PD, Port SA, Patena W, et al. The structural basis of rubisco phase separation in the pyrenoid. Nat Plants. 2020:6(12):1480–1490. 10.1038/s41477-020-00811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann G, Mahboubi A, Bente H, Garcia D, Hanson J, Hafr NA. Arabidopsis RNA processing body components LSM1 and DCP5 aid in the evasion of translational repression during Cauliflower mosaic virus infection. Plant Cell. 2022:34(8):3128–3147. 10.1093/plcell/koac132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yang M, Zhang X. The function of small RNAs in plant biotic stress response. J Integr Plant Biol. 2016:58(4):312–327. 10.1111/jipb.12463 [DOI] [PubMed] [Google Scholar]
- Huang S, Zhu S, Kumar P, MacMicking JD. A phase-separated nuclear GBPL circuit controls immunity in plants. Nature. 2021:594(7863):424–429. 10.1038/s41586-021-03572-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-S, Wei T, Laliberté J-F, Wang A. A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 2010:152(1):255–266. 10.1104/pp.109.147983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014:30(1):39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Ismail H, Liu X, Yang F, Li J, Zahid A, Dou Z, Liu X, Yao X. Mechanisms and regulation underlying membraneless organelle plasticity control. J. Mol Cell Biol. 2021:13(4):239–258. 10.1093/jmcb/mjab028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G-J, Jang J-C, Wu S-H. Dynamics and functions of stress granules and processing bodies in plants. Plants (Basel). 2020:9(9):1122. 10.3390/plants9091122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang GJ, Yang JY, Hsieh HL, Wu SH. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc Natl Acad Sci U S A. 2019:116(13):6451–6456. 10.1073/pnas.1900084116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T, Zhang B, You C, Zhang Y, Zeng L, Li S, Johnson KCM, Yu B, Li X, Chen X. The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell. 2017:29(10):2626–2643. 10.1105/tpc.17.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Korasick DA, Emenecker RJ, Morffy N, Wilkinson EG, Powers SK, Strader LC. Regulation of AUXIN RESPONSE FACTOR condensation and nucleo-cytoplasmic partitioning. Nat Commun. 2022:13(1):4015. 10.1038/s41467-022-31628-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012:31(7):1704–1713. 10.1038/emboj.2012.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020:585(7824):256–260. 10.1038/s41586-020-2644-7 [DOI] [PubMed] [Google Scholar]
- Kearly A, Nelson ADL, Skirycz A, Chodasiewicz M. Composition and function of stress granules and P-bodies in plants. Semin Cell Dev Biol. 2022:S1084-9521(22):00350-0. 10.1016/j.semcdb.2022.11.008 [DOI] [PubMed] [Google Scholar]
- Kim EY, Wang L, Lei Z, Li H, Fan W, Cho J. Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nat Plants. 2021:7(3):303–309. 10.1038/s41477-021-00867-4 [DOI] [PubMed] [Google Scholar]
- Kim SH, Macfarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, Haupt S, Brown JW, Taliansky M. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc Natl Acad Sci U S A. 2007a:104(26):11115–11120. 10.1073/pnas.0704632104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JW, Taliansky M. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007b:26(8):2169–2179. 10.1038/sj.emboj.7601674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ. Crosstalk between RNA silencing and RNA quality control in plants. BMB Rep. 2023:56(6):321–325. 10.5483/BMBRep.2023-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. The role of mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011:30(5):814–822. 10.1038/emboj.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop K, Stepien A, Barciszewska-Pacak M, Taube M, Bielewicz D, Michalak M, Borst JW, Jarmolowski A, Szweykowska-Kulinska Z. Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. 2017:45(5):2757–2775. 10.1093/nar/gkw895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan C, Strader LC, Morffy N. ARF19 Condensation in the Arabidopsis stomatal lineage. MicroPubl Biol. 2023:2023:10.17912/micropub.biology.000708. 10.17912/micropub.biology.000708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 And RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009:583(8):1261–1266. 10.1016/j.febslet.2009.03.055 [DOI] [PubMed] [Google Scholar]
- Kunej U, Jakse J, Radisek S, Stajner N. Core RNA interference genes involved in miRNA and ta-siRNA biogenesis in hops and their expression analysis after challenging with verticillium nonalfalfae. Int J Mol Sci. 2021:22(8):4224. 10.3390/ijms22084224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Wang L, Kim EY, Cho J. Phase separation of chromatin and small RNA pathways in plants. Plant J. 2021:108(5):1256–1265. 10.1111/tpj.15517 [DOI] [PubMed] [Google Scholar]
- Li CF, Henderson IR, Song L, Fedoroff N, Lagrange T, Jacobsen SE. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet. 2008:4(2):e27. 10.1371/journal.pgen.0040027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006:126(1):93–106. 10.1016/j.cell.2006.05.032 [DOI] [PubMed] [Google Scholar]
- Li F, Huang C, Li Z, Zhou X. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 2014:10(2):e1003921. 10.1371/journal.ppat.1003921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhao N, Li Z, Xu X, Wang Y, Yang X, Liu SS, Wang A, Zhou X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017:13(2):e1006213. 10.1371/journal.ppat.1006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu H, Liu K, Yang W, Zhou B, Gan L, Li S, Zhang C, Yu B. Serrate-associated protein 1, a splicing-related protein, promotes miRNA biogenesis in Arabidopsis. New Phytol. 2021a:232(5):1959–1973. 10.1111/nph.17691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liu N, Liu Q, Zheng X, Lu L, Gao W, Liu Y, Liu Y, Zhang S, Wang Q, et al. DEAD-box helicases modulate dicing body formation in Arabidopsis. Sci Adv. 2021b:7(18):eabc6266. 10.1126/sciadv.abc6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li M, Liu K, Zhang H, Zhang S, Zhang C, Yu B. MAC5, An RNA-binding protein, protects pri-miRNAs from SERRATE-dependent exoribonuclease activities. Proc Natl Acad Sci U S A. 2020:117(38):23982–23990. 10.1073/pnas.2008283117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu K, Zhou B, Li M, Zhang S, Zeng L, Zhang C, Yu B. MAC3A And MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 2018a:30(2):481–494. 10.1105/tpc.17.00953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013:153(3):562–574. 10.1016/j.cell.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu R, Li A, Liu K, Gu L, Li M, Zhang H, Zhang Y, Zhuang S, Wang Q, et al. SMA1, A homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2018b:46(17):9148–9159. 10.1093/nar/gky591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q, Fang Y, Yu Y, Lv H, Zheng Y, et al. Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet. 2016:12(11):e1006422. 10.1371/journal.pgen.1006422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Fang X. Phase separation in RNA biology. J Genet Genomics. 2021:48(10):872–880. 10.1016/j.jgg.2021.07.012 [DOI] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009:20(6):1661–1670. 10.1091/mbc.e08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen X. RNA Quality control as a key to suppressing RNA silencing of endogenous genes in plants. Mol Plant. 2016:9(6):826–836. 10.1016/j.molp.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gao W, Wu S, Lu L, Chen Y, Guo J, Men S, Zhang X. AtXRN4 affects the turnover of chosen miRNA*s in Arabidopsis. Plants (Basel). 2020:9(3):362. 10.3390/plants9030362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love AJ, Yu C, Petukhova NV, Kalinina NO, Chen J, Taliansky ME. Cajal bodies and their role in plant stress and disease responses. RNA Biol. 2017:14(6):779–790. 10.1080/15476286.2016.1243650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Nicole MC, Meteignier LV, Hong N, Wang G, Moffett P. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J Exp Bot. 2015:66(3):919–932. 10.1093/jxb/eru447 [DOI] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA. 2013:4(1):17–34. 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- Makarov V, Rakitina D, Protopopova A, Yaminsky I, Arutiunian A, Love AJ, Taliansky M, Kalinina N. Plant coilin: structural characteristics and RNA-binding properties. PLoS One. 2013:8(1):e53571. 10.1371/journal.pone.0053571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen K, Lohmus A, Pollari M. Plant RNA regulatory network and RNA granules in virus infection. Front Plant Sci. 2017:8:2093. 10.3389/fpls.2017.02093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD. Composition and function of P bodies in Arabidopsis thaliana. Front Plant Sci. 2014:5:201. 10.3389/fpls.2014.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS. Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerg Top Life Sci. 2020:4(3):307–329. 10.1042/ETLS20190164 [DOI] [PubMed] [Google Scholar]
- Martinez de Alba AE, Moreno AB, Gabriel M, Mallory AC, Christ A, Bounon R, Balzergue S, Aubourg S, Gautheret D, Crespi MD, et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015:43(5):2902–2913. 10.1093/nar/gkv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navascues J, Bengoechea R, Tapia O, Casafont I, Berciano MT, Lafarga M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J Struct Biol. 2008:163(2):137–146. 10.1016/j.jsb.2008.04.013 [DOI] [PubMed] [Google Scholar]
- Okano Y, Senshu H, Hashimoto M, Neriya Y, Netsu O, Minato N, Yoshida T, Maejima K, Oshima K, Komatsu K, et al. In planta recognition of a double-stranded RNA synthesis protein complex by a potexviral RNA silencing suppressor. Plant Cell. 2014:26(5):2168–2183. 10.1105/tpc.113.120535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Li X, Zhang J, Feng P, Pu H, Kong L, Bai Z, Rong L, Xu X, Chi W, et al. Liquid-liquid phase transition drives intra-chloroplast cargo sorting. Cell. 2020:180(6):1144–1159.e20. 10.1016/j.cell.2020.02.045 [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000:151(7):1561–1574. 10.1083/jcb.151.7.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006:126(1):79–92. 10.1016/j.cell.2006.05.031 [DOI] [PubMed] [Google Scholar]
- Pyo Y, Kim GM, Choi SW, Song CY, Yang SW, Jung IL. Strontium stress disrupts miRNA biogenesis by reducing HYL1 protein levels in Arabidopsis. Ecotoxicol Environ Saf. 2020:204:111056. 10.1016/j.ecoenv.2020.111056 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Shi J, Zhai Y, Hou Y, Ma W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci U S A. 2015:112(18):5850–5855. 10.1073/pnas.1421475112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamaki ML, Streng J, Valkonen JP. Silencing suppressor protein VPg of a potyvirus interacts with the plant silencing-related protein SGS3. Mol Plant Microbe Interact. 2014:27(11):1199–1210. 10.1094/MPMI-04-14-0109-R [DOI] [PubMed] [Google Scholar]
- Rajamaki ML, Valkonen JP. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell. 2009:21(8):2485–2502. 10.1105/tpc.108.064147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008:321(5895):1490–1492. 10.1126/science.1163728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A. 2012:109(31):12817–12821. 10.1073/pnas.1204915109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci U S A. 2014:111(17):6365–6370. 10.1073/pnas.1405083111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhine K, Vidaurre V, Myong S. RNA droplets. Annu Rev Biophys. 2020:49(1):247–265. 10.1146/annurev-biophys-052118-115508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ruiz S, Soler N, Sanchez-Navarro J, Fagoaga C, Lopez C, Navarro L, Moreno P, Pena L, Flores R. Citrus tristeza virus p23: determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis. Mol Plant Microbe Interact. 2013:26(3):306–318. 10.1094/MPMI-08-12-0201-R [DOI] [PubMed] [Google Scholar]
- Semashko MA, Rakitina DV, Gonzalez I, Canto T, Kalinina NO, Taliansky ME. Movement protein of hordeivirus interacts in vitro and in vivo with coilin, a major structural protein of Cajal bodies. Dokl Biochem Biophys. 2012:442(1):57–60. 10.1134/S1607672912010164 [DOI] [PubMed] [Google Scholar]
- Shang B, Wang L, Yan X, Li Y, Li C, Wu C, Wang T, Guo X, Choi SW, Zhang T, et al. Intrinsically disordered proteins SAID1/2 condensate on SERRATE for dual inhibition of miRNA biogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2023:120(14):e2216006120. 10.1073/pnas.2216006120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Yu C, Makhotenko AV, Makarova SS, Love AJ, Kalinina NO, MacFarlane S, Chen J, Taliansky ME. Interaction of a plant virus protein with the signature Cajal body protein coilin facilitates salicylic acid-mediated plant defence responses. New Phytol. 2019:224(1):439–453. 10.1111/nph.15994 [DOI] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, MacRae IJ. Phase transitions in the assembly and function of human miRISC. Cell. 2018:173(4):946–957. 10.1016/j.cell.2018.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017:357(6357):eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Rajeswaran R, Blevins T, Schoelz J, Meins F Jr, Hohn T, Pooggin MM. The CaMV transactivator/viroplasmin interferes with RDR6-dependent trans-acting and secondary siRNA pathways in Arabidopsis. Nucleic Acids Res. 2008:36(18):5896–5909. 10.1093/nar/gkn590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink JAFO, Loayza-Puch F, Elkon R, Agami R. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017:169(2):326–337.e12. 10.1016/j.cell.2017.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Miranda J, Chodasiewicz M, Skirycz A, Fernie AR, Moschou PN, Bozhkov PV, Gutierrez-Beltran E. Stress-related biomolecular condensates in plants. Plant Cell. 2023:35(9):3187–3204. 10.1093/plcell/koad127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A. 2007:104(13):5437–5442. 10.1073/pnas.0701061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Yang J, Wang C, Lu Q, Shi L, Tayier S, Jia G. Arabidopsis N(6)-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol Plant. 2021:14(4):571–587. 10.1016/j.molp.2021.01.014 [DOI] [PubMed] [Google Scholar]
- Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol. 2019:70(1):489–525. 10.1146/annurev-arplant-050718-100334 [DOI] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004:15(2):173–183. 10.1016/j.molcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Stepien A, Dolata J, Gulanicz T, Bielewicz D, Bajczyk M, Smolinski DJ, Szweykowska-Kulinska Z, Jarmolowski A. Chromatin-associated microprocessor assembly is regulated by the U1 snRNP auxiliary protein PRP40. Plant Cell. 2022:34(12):4920–4935. 10.1093/plcell/koac278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Li Z, Cheng J, Li L, Zhong S, Liu L, Zheng Y, Zheng B. The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev Cell. 2017:41(5):527–539.e5. 10.1016/j.devcel.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Sun Z, Guo T, Liu Y, Liu Q, Fang Y. The roles of Arabidopsis CDF2 in transcriptional and posttranscriptional regulation of primary microRNAs. PLoS Genet. 2015:11(10):e1005598. 10.1371/journal.pgen.1005598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Li M, Zhou Y, Guo T, Liu Y, Zhang H, Fang Y. Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLoS Genet. 2018:14(3):e1007247. 10.1371/journal.pgen.1007247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliansky M, Love AJ, Kolowerzo-Lubnau A, Smolinski DJ. Cajal bodies: evolutionarily conserved nuclear biomolecular condensates with properties unique to plants. Plant Cell. 2023:35(9):3214–3235. 10.1093/plcell/koad140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Luo W, Yan W, Liu J, Aizezi Y, Cui R, Tian R, Ma J, Guo H. Phase separation of SGS3 drives siRNA body formation and promotes endogenous gene silencing. Cell Rep. 2023:42(1):111985. 10.1016/j.celrep.2022.111985 [DOI] [PubMed] [Google Scholar]
- Thran M, Link K, Sonnewald U. The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 2012:72(3):368–377. 10.1111/j.1365-313X.2012.05066.x [DOI] [PubMed] [Google Scholar]
- Tibble RW, Gross JD. A call to order: examining structured domains in biomolecular condensates. J Magn Reson. 2023:346:107318. 10.1016/j.jmr.2022.107318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassi AH, Re DA, Romani F, Cambiagno DA, Gonzalo L, Moreno JE, Arce AL, Manavella PA. The intrinsically disordered protein CARP9 bridges HYL1 to AGO1 in the nucleus to promote microRNA activity. Plant Physiol. 2020:184(1):316–329. 10.1104/pp.20.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Ren Z, Sun L, Zhou S, Yuan W, Hui Y, Ci D, Wang W, Fan LM, Wu Z, et al. ALBA Proteins confer thermotolerance through stabilizing HSF messenger RNAs in cytoplasmic granules. Nat Plants. 2022:8(7):778–791. 10.1038/s41477-022-01175-1 [DOI] [PubMed] [Google Scholar]
- Tong X, Liu SY, Zou JZ, Zhao JJ, Zhu FF, Chai LX, Wang Y, Han C, Wang XB. A small peptide inhibits siRNA amplification in plants by mediating autophagic degradation of SGS3/RDR6 bodies. EMBO J. 2021:40(15):e108050. 10.15252/embj.2021108050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, Zhao Y, Yu Y, Ramachandran V, Ren G, et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 2015:11(4):e1005119. 10.1371/journal.pgen.1005119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the Cajal body marker protein, coilin. PLoS One. 2009:4(7):e6171. 10.1371/journal.pone.0006171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang H, Huai J, Peng F, Wu J, Lin R, Fang X. Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat Chem Biol. 2022:18(12):1361–1369. 10.1038/s41589-022-01196-z [DOI] [PubMed] [Google Scholar]
- Wang J, Chen S, Jiang N, Li N, Wang X, Li Z, Li X, Liu H, Li L, Yang Y, et al. Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana. Nucleic Acids Res. 2019a:47(15):7886–7900. 10.1093/nar/gkz526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ding Y, He L, Zhang G, Zhu JK, Lozano-Duran R. A virus-encoded protein suppresses methylation of the viral genome through its interaction with AGO4 in the Cajal body. Elife. 2020:9:e55542. 10.7554/eLife.55542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Song X, Gu L, Li X, Cao S, Chu C, Cui X, Chen X, Cao X. NOT2 Proteins promote polymerase II-dependent transcription and interact with multiple microRNA biogenesis factors in Arabidopsis. Plant Cell. 2013:25(2):715–727. 10.1105/tpc.112.105882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Quan L, Li S, You C, Zhang Y, Gao L, Zeng L, Liu L, Qi Y, Mo B, et al. The PROTEIN PHOSPHATASE4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell. 2019b:31(2):486–501. 10.1105/tpc.18.00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Gu Y. The emerging role of biomolecular condensates in plant immunity. Plant Cell. 2022:34(5):1568–1572. 10.1093/plcell/koab240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y. An importin beta protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell. 2011:23(10):3565–3576. 10.1105/tpc.111.091058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Dou Y, Chen L, Wang J, Jiang N, Guo C, Yao Q, Wang C, Liu L, et al. Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3′ to 5′ exoribonuclease atrimmer 2 in Arabidopsis. Proc Natl Acad Sci U S A. 2018a:115(28):E6659–E6667. 10.1073/pnas.1721917115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang S, Dou Y, Zhang C, Chen X, Yu B, Ren G. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet. 2015:11(4):e1005091. 10.1371/journal.pgen.1005091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma Z, Castillo-Gonzalez C, Sun D, Li Y, Yu B, Zhao B, Li P, Zhang X. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature. 2018b:557(7706):516–521. 10.1038/s41586-018-0135-x [DOI] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008:56(4):517–530. 10.1111/j.1365-313X.2008.03623.x [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009:136(3):496–507. 10.1016/j.cell.2008.12.023 [DOI] [PubMed] [Google Scholar]
- Wu X, Shi Y, Li J, Xu L, Fang Y, Li X, Qi Y. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013:23(5):645–657. 10.1038/cr.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T, Cheng SLH, Lai SK, Li HY, Mueller-Cajar O. The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat Commun. 2018:9(1):5076. 10.1038/s41467-018-07624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Chen M, Niu J, Wang L, Li Y, Fang X, Li P, Qi Y. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat Cell Biol. 2021:23(1):32–39. 10.1038/s41556-020-00606-5 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005:138(4):2145–2154. 10.1104/pp.105.062943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zheng C, Lu D, Song CP, Zhang L. Phase separation in plants: new insights into cellular compartmentalization. J Integr Plant Biol. 2021:63(11):1835–1855. 10.1111/jipb.13152 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang P, Wang B, Hsu CC, Tang K, Zhang H, Hou YJ, Zhao Y, Wang Q, Zhao C, et al. The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 2017:13(4):e1006753. 10.1371/journal.pgen.1006753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc Natl Acad Sci U S A. 2012:109(1):315–320. 10.1073/pnas.1114673109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Yang J, Sun Y, Zhao P, Gao S, Jung C, Qu J, Fang R, Chua NH. Geminivirus activates ASYMMETRIC LEAVES 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLoS Pathog. 2015:11(10):e1005196. 10.1371/journal.ppat.1005196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008:105(29):10073–10078. 10.1073/pnas.0804218105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ji L, Le BH, Zhai J, Chen J, Luscher E, Gao L, Liu C, Cao X, Mo B, et al. ARGONAUTE10 Promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017:15(2):e2001272. 10.1371/journal.pbio.2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci U S A. 2012:109(44):18198–18203. 10.1073/pnas.1216199109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, You C, Zhang Y, Zeng L, Hu J, Zhao M, Chen X. Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis. Nat Plants. 2020:6(8):957–969. 10.1038/s41477-020-0726-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Yu B. PRL1, An RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 2014:10(12):e1004841. 10.1371/journal.pgen.1004841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xie M, Ren G, Yu B. CDC5, A DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc Natl Acad Sci U S A. 2013:110(43):17588–17593. 10.1073/pnas.1310644110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Y, Liu X, Hong X, Xu Y, Zhu P, Shen Y, Wu H, Ji Y, Wen X, et al. Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015:348(6230):120–123. 10.1126/science.aaa2618 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan S, Hua C, Teo ZWN, Kiang JX, Shen L, Yu H. Phase separation of HRLP regulates flowering time in Arabidopsis. Sci Adv. 2022:8(25):eabn5488. 10.1126/sciadv.abn5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Niu J, Hai Z, Li T, Xie D, Li Y, Qi Y. Peptidyl-prolyl isomerase Cyclophilin71 promotes SERRATE phase separation and miRNA processing in Arabidopsis. Proc Natl Acad Sci U S A. 2023:120(36):e2305244120. 10.1073/pnas.2305244120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Thanh Theresa D, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012:22(8):689–694. 10.1016/j.cub.2012.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Zhang H. Phase separation in membrane biology: the interplay between membrane-bound organelles and membraneless condensates. Dev Cell. 2020:55(1):30–44. 10.1016/j.devcel.2020.06.033 [DOI] [PubMed] [Google Scholar]
- Zheng L, He J, Ding Z, Zhang C, Meng R. Identification of functional domain(s) of fibrillarin interacted with p2 of Rice stripe virus. Can J Infect Dis Med Microbiol. 2018:2018:8402839. 10.1155/2018/8402839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Gu J, Yao J, Li Y, Zhang Z, Xia W, Wang Z, Gui X, Li L, Li D, et al. Liquid-liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev Cell. 2022:57(5):583–597.e6. 10.1016/j.devcel.2022.02.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors of distribution of materials integral to findings in this article according to the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Xiangming Zhang (zhangxm@ioz.ac.cn), Qi Li (liqi@ioz.ac.cn) and Yang Liu (liuyang2016@ioz.ac.cn). All data are incorporated into the article and its online supplementary material.