Abstract
The innate immune system protects the host from external pathogens and internal damage in various ways. The cGAS-STING signaling pathway, comprised of cyclic GMP-AMP synthase (cGAS), stimulator of interferon genes (STING), and downstream signaling adaptors, plays an essential role in protective immune defense against microbial DNA and internal damaged-associated DNA and is responsible for various immune-related diseases. After binding with DNA, cytosolic cGAS undergoes conformational change and DNA-linked liquid-liquid phase separation to produce 2'3'-cGAMP for the activation of endoplasmic reticulum (ER)-localized STING. However, further studies revealed that cGAS is predominantly expressed in the nucleus and strictly tethered to chromatin to prevent binding with nuclear DNA, and functions differently from cytosolic-localized cGAS. Detailed delineation of this pathway, including its structure, signaling, and regulatory mechanisms, is of great significance to fully understand the diversity of cGAS-STING activation and signaling and will be of benefit for the treatment of inflammatory diseases and cancer. Here, we review recent progress on the above-mentioned perspectives of the cGAS-STING signaling pathway and discuss new avenues for further study.
Keywords: cGAS, STING, Structure, Signaling, Post-translational modification, Diseases
INTRODUCTION
Pattern recognition receptors (PRRs) mediate cell perception of extracellular pathogen infection and intracellular damage signals (Tao et al., 2016). PRRs recognize pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) to activate downstream signaling pathways and mount effective innate immune responses. For example, Toll-like receptor 3 (TLR3), TLR7, TLR8, TLR9, and TLR13 recognize intracellular pathogenic nucleic acids (Broz & Dixit, 2016), retinoic acid-inducible gene I (RIG-I)-like receptors recognize viral RNA (Yoneyama et al., 2015), NOD-like receptors detect components of bacterial cell walls (Kaparakis et al., 2007), and cyclic GMP-AMP synthase (cGAS) recognizes exogenous or endogenous leaked DNA to launch immune defenses.
First discovered as a cytosolic-resident DNA sensor for non-self-DNA and damage-associated self-DNA, cGAS catalyzes the production of second messenger 2′3′-cGAMP from adenosine triphosphate (ATP) and guanosine triphosphate (GTP) upon DNA binding. This second messenger activates an interferon (IFN)-stimulated protein (stimulator of interferon genes, STING) localized in the endoplasmic reticulum (ER), which, upon activation, induces the production of type I IFN (IFN-I) via IFN regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB) activation, protecting the host from infection and initiating antiviral and antitumor innate immune responses (Burdette et al., 2011). However, later studies revealed that cGAS is actually predominantly expressed in the nucleus and is tethered tightly with chromatin (Chen et al., 2020; Liu et al., 2018a; Volkman et al., 2019), raising questions of how cGAS is regulated to prevent its recognition of nuclear DNA, what is the main function of nuclear-located cGAS, and are there any structural discrepancies between cytosolic and nuclear-localized cGAS? Activation and deactivation of cGAS and STING are tightly regulated to ensure proper initiation of the immune response and maintenance of host homeostasis, and cytosolic and nuclear cGAS may adopt different regulation mechanisms due to differences in cellular regions and interactions with different proteins. As a signaling pathway responsible for the pathogenesis of many diseases, the role of cGAS-STING signaling in disease control and drug development (e.g., inhibitors of cGAS and STING and cGAS and STING drugs in combination with other drugs to overcome resistance) has become a focus for many researchers (Decout et al., 2021).
Here, we review the structural and functional basis of cGAS (cytosolic and nuclear) and STING, discuss the activation models and regulatory mechanisms underlying cytosolic and nuclear cGAS discrepancies and STING activation, and explore the role of cGAS-STING signaling in disease pathogenesis and treatment. We also discuss current limitations and future avenues for research on this signaling pathway.
STRUCTURAL BASIS OF CGAS-STING-MEDIATED DNA RECOGNITION AND SIGNAL TRANSDUCTION
Structure of cGAS
Since its purification and identification in 2012 (Sun et al., 2013), research on the structure and function of cGAS has become a hotspot in the immunology field. The cGAS protein belongs to the nucleotidyl transferase (NTase) superfamily, which can recognize exogenously or endogenously released nucleic acids (Sun et al., 2013). Structurally, cGAS consists of an N-terminal positively charged domain responsible for cGAS dimer stabilization and a globular C-terminal catalytic domain consisting of a NTase core for enzymatic activity and Mab21 domain for DNA binding. The Mab21 domain is structurally comprised of two lobes divided by a deep cleft required for nucleotide binding. The N-terminal lobe is composed of β-sheets, flanked by α-helices on the side, while the C-terminal lobe is a helix bundle with a conserved zinc (Zn) thumb. The catalytic residues of cGAS are located on the central β-sheet of the NTase domain, with their side chains pointing to the cleft that separates the N and C lobes (Civril et al., 2013; Zhang et al., 2014). Mutations of catalytic residues Glu200 and Asp202 into Gln and Asn, respectively, diminish catalytic activity in porcine cGAS (pcGAS) (Kranzusch et al., 2013). Mutations of Gly212, Ser213, Glu225, Asp227, and Asp319 into alanine also result in a loss of catalytic activity in human cGAS (hcGAS) (Kranzusch et al., 2013; Li et al., 2013). The conserved Zn thumb is important for cGAS dimerization and DNA binding (Laity et al., 2001). The H(X5)CC(X6)C Zn-binding motif inserted between residues 389 and 405 induces structural rearrangement of the cGAS C-terminal domain upon binding to Zn. These Zn ligand sites support charged loops that alter the gap required for DNA binding (Kranzusch et al., 2013).
Structure of cGAS-DNA interactions
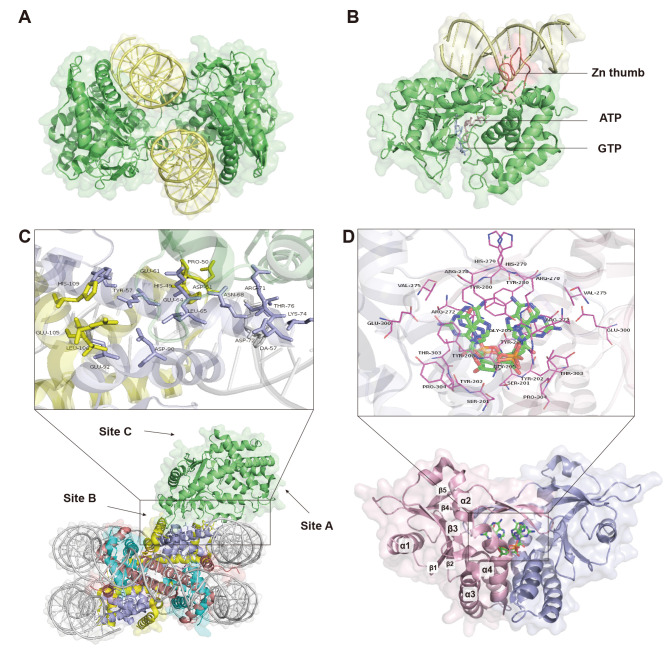
Cytoplasmic cGAS-DNA complex: After binding to appropriate length DNA, cGAS undergoes structural transformation for dimer assembly (Civril et al., 2013). Two double-stranded DNA (dsDNA) molecules and two cGAS molecules form a minimal 2:2 complex, with one DNA molecule binding to one cGAS molecule and the two strands sandwiched in the middle of the complex (Figure 1A). Interactions between the two cGAS molecules are mediated by the 15 bases of the backbone chain and the hydrogen bonds of the side chain. Residues Asn389 to Glu398 in the ring of the Zn thumb contribute to dimer formation (Gao et al., 2013b; Li et al., 2013). Residues Lys347 (Lys335 of mouse cGAS (mcGAS)) and Lys394 (Lys382 of mcGAS) of hcGAS mediate dimer formation (Li et al., 2013; Zhang et al., 2014). Although the binding of cGAS to DNA is independent of its sequence, there is a requirement for DNA length. Theoretically, DNA lengths of 16–17 bp are sufficient to activate cGAS (Zhou et al., 2019), but DNA fragments >45 bp are required for optimal cGAS recognition and cGAMP production. According to recent studies, long DNA is recognized by the construction of higher order cGAS-DNA “ladder” complexes, in which DNA-binding proteins are stabilized by pre-arranging DNA into a U-shape, with multiple cGAS dimers binding to the DNA ladder in succession. This structure also suggests that protein-DNA interactions are required in the context of long DNA recognition (Andreeva et al., 2017). Binding of DNA to the positively charged N-terminus of cGAS can stably induce the generation of liquid-like droplets and enhance the enzymatic activity of cGAS by mediating liquid phase condensation (Xie et al., 2019). In particular, long DNA-cGAS complexes can achieve higher-order oligomerization by increasing cGAS concentration, facilitating cGAS liquid phase separation (Du & Chen, 2018), and mediating stable cGAS activation (Zhou et al., 2018).
Figure 1.
Structural basis of cGAS-STING-mediated DNA recognition and signal transduction
A: Two dsDNA molecules and two cGAS molecules form a minimal 2:2 complex. Structure is constructed according to PDB: 4O6A, adapted from Zhang et al. (2014) with slight modifications. cGAS is in green, DNA is in yellow. B: Structure of cytoplasmic cGAS binding to DNA constructed according to PDB: 4KB6, adapted from Civril et al. (2013) with slight modifications. Zn thumb domain is in red, ATP is in light pink, and GTP is in light blue. C: Structure of cGAS nucleosome complex constructed according to PDB: 6Y5E, adapted from Pathare et al. (2020) with slight modifications. cGAS is in green, black box shows details of cGAS and nucleosome counteraction, sites A, B, and C are cGAS-DNA binding sites. D: Structure of STING binding with 2′3′-cGAMP constructed according to PDB: 5CFQ, adapted from Kranzusch et al. (2013) with slight modifications. Black boxes show details of contact between 2′3′-cGAMP and STING. 2′3′-cGAMP is in blue-green sticks. STING dimers are ribbon structures, with monomers in pink and purple, α helices and β folding are labeled.
Nuclear cGAS-DNA complex: Nuclear-localized cGAS exhibits distinct functions and DNA binding from cytoplasmic cGAS. Intranuclear cGAS can inhibit DNA repair and attenuate DNA replication (Bai & Liu, 2022). Although its binding affinity to endogenous DNA is 200-fold lower than its exogenous counterpart (Gentili et al., 2019), self-DNA sensing in the nucleus is still tightly controlled to avoid unnecessary activation of the immune response, which mainly relies on the interaction structures.
During the interphase of a cell cycle, DNA-protein complexes in the nucleus exist primarily as chromatin. Chromatin is assembled by nucleosome core particles, which are composed of approximately 145 bp of DNA and histones H2A, H2B, H3, and H4 (Min & Liu, 2021; Parmar & Padinhateeri, 2020). Similar to the cGAS-DNA complex in the cytoplasm, the cGAS monomer in the nucleus bind to a nucleosome, and by interacting with nucleosomal DNA of another cGAS-nucleosome complex, two cGAS molecules are sandwiched between two nucleosomes to form a 2:2 complex (Cao et al., 2020). However, unlike cytoplasmic cGAS dimers, cGAS in the cGAS-nucleosome 2:2 complex is not dimerized.
Nuclear cGAS has three DNA-binding sites (sites A, B, and C; Figure 1C) (Michalski et al., 2020). Firstly, cGAS interacts with the histone H2A–H2B heterodimer around the B site, and the side chains of Arg222, Lys240, and Arg241 of mcGAS interact with the carboxyl groups of Glu61, Glu64, Asp90, and Glu92 of histone H2A through electrostatic interactions, anchoring cGAS to the histone H2A–H2B heterodimer acidic patch and preventing DNA from entering the B site (Pathare et al., 2020; Zhao et al., 2020). Secondly, nucleosomal DNA in superhelix position 6 or 7 (SHL6 or SHL7) occupies the region of the second cGAS molecule in the cGAS dimer. Thus, nucleosome binding spatially blocks cGAS dimer formation, even if the nucleosomal DNA binds to sites A and C, as binding to DNA at sites A and B as well as dimer formation are essential for cGAS activation (Cao et al., 2020; Michalski et al., 2020). In this way, the chromatin is tightly tethered to cGAS, preventing it from reacting with self-DNA.
Structural basis of 2'3'-cGAMP production
STING primarily binds to and induces IFN-I production from two cyclic dinucleotides, c-di-AMP and c-di-GMP, identified as second messengers secreted by bacteria (Sauer et al., 2011; Yin et al., 2012). The Vibrio seventh pandemic island-1 (VSP-1) gene encodes a class of dinucleotide cyclases that preferentially synthesize cyclic-GMP-AMP molecules, leading to speculation of whether new cyclic dinucleotide species are also capable of activating STING (Davies et al., 2012). Previous studies have shown that cGAS catalyzes cGAMP production from ATP and GTP in the presence of DNA (Wu et al., 2013). This cGAMP was later identified as 2'3'-cGAMP (Gao et al., 2013b), which binds to and activates STING as a second messenger. These findings suggest that 2'3'-cGAMP is an endogenous second messenger generated by cGAS in mammalian cells.
Upon binding to dsDNA, cGAS recruits ATP and GTP (Figure 1B). First, cGAS, ATP, and dsDNA form a ternary complex, where ATP first binds in a catalytic pocket inside cGAS. The side chain of Glu211 in cGAS travels towards the other two acidic residues of the ternary complex. The phosphate group of ATP triphosphate moiety binds to the polar side chains of cGAS (Ser199, Ser420, and Lys402), acting as a bridge between the two binding cations (Mg2+). GTP then forms a phosphodiester bond in the catalytic pocket of cGAS, resulting in the bound ligand 5'-pppGpG. Surprisingly, the GpG linkage of 5'-pppGpG is 2',5' rather than the commonly assumed 3',5', and the first and second G residues adopt cis and antiglycosidic reversal orientations. The triphosphate group of 5'-pppG(2',5')pG is coordinated to two cations. The first G stacks on Tyr421 and the second G uses its Watson-Crick edge to form hydrogen bonds with the polar side chains (Thr197, Ser366, and Ser368) and its Hoogsteen edge to form a hydrogen bond with Arg364. If cGAS, ATP, and GTP are stacked together, the first G binds to the ATP plane (Gao et al., 2013b). Then, cGAS catalyzes the synthesis of linear 2'-5'-linked dinucleotides, then synthesizes 3'-5' phosphodiester bonds between AMP of 3'-OH and GMP of 5'-phosphate through flip-flop conversion in the catalytic pocket to obtain the cyclic end-product. The dissociation constants (Kd) of 2'3'-cGAMP, 3'3'-cGAMP, and 2'2'-cGAMP are higher than that of 2'3'-cGAMP, and thus 2'3'-cGAMP exhibits the highest affinity for STING (Shi et al., 2015; Yin et al., 2012).
Structure of STING and 2'3'-cGAMP binding
STING is a transmembrane protein located in the ER, comprising a transmembrane (TM) domain with four TM helices, a cytoplasmic ligand-binding domain (LBD) essential for dimerization and cGAMP binding, and a C-terminal tail (CTT) containing the TANK-binding kinase 1 (TBK1) phosphorylation site required for downstream signaling (Shang et al., 2019; Tsuchida et al., 2010). When its function was first demonstrated, STING was considered an adaptor molecule essential for IFN-I induction by non-intracellular DNA produced by various DNA pathogens, e.g., herpes simplex virus (HSV-1) (Sun et al., 2009), after infection (Ishikawa & Barber, 2008). Further studies have validated the ability of STING to produce IFN-I via TBK1 and transcription factor IRF3 (Scheres, 2012; Sun et al., 2009).
The crystal structure of the STING cytoplasmic domain is a highly structured butterfly-like hydrophobic dimer (Ergun & Li, 2020). The two LBDs form a ligand-binding pocket in a deep V-shaped cleft between the dimer subunits (Shang et al., 2019). The STING dimer, similar to its overall mono-structure, is characterized by five twisted β-sheets surrounded by four α-helices (Cong et al., 2019). In the STING dimer, TM1 of one STING molecule and TM2, TM3, and TM4 of another molecule are packed together to form a domain-wrapped architecture. The eight TM helices in the STING dimer are organized into two layers: i.e., middle layer formed by TM2 and TM4 from both subunits and periphery formed by TM1 and TM3. TM2 (residues 45–69), the longest TM helix in STING, is substantially tilted relative to the membrane plane. A two-turn amphipathic connector helix exists between TM4 and the first helix of the LBD. The two connector helices in the dimer form a tiny right-handed coil on the cytoplasmic surface of the TM domain, passing through the hydrophobic surface (Shang et al., 2019). Two short helices form the linker between TM2 and TM3 (residues 70–91), with the C-terminus of the second helix strongly attached to the N-terminus of the aforementioned connector helix. Between the cytoplasmic and TM domains, the TM2–TM3 linker, connector helix, and LBD form a surface groove. The N-terminal portion (residues 4–15) lies snugly in the groove preceding TM1. This conserved structure is critical for the stability and function of the protein.
STING can be activated by 2'3'-cGAMP (hereinafter referred to as cGAMP) and c-di-GMP (Burdette et al., 2011; Huang et al., 2012; Ouyang et al., 2012). Here, we focus on the structure of cGAMP when combined with STING. Bound cGAMP molecules are positioned on the two-fold axis of the butterfly-like dimer (Figure 1D). cGAMP adopts two orientations associated with the double symmetry, compatible with the two initial oblique junctions in the STING dimer bound to guanine or adenine. Due to its asymmetric nature, cGAMP can bind to two symmetric STING dimers with opposite orientations (Shi et al., 2015).
cGAMP binds to the ligand-binding pocket of the STING dimer, which is uncharged at its base, but has positively and negatively charged residues arranged on its walls, and interdomain interactions involving several pairs of polar contacts. Interdomain interactions include interactions between the side group of Tyr245 and main chain carbonyl oxygen atom of Gly234, between the side group of Ser243 and main chain amide nitrogen atom of Lys236, and between the side groups of Asp237 and Lys224. The bound cGAMP is anchored by purine bases on either side by Tyr167 and Arg238 of STING, which are arranged in a plane to form a hydrogen bond with Asn7 of one purine and a guanidine group stack on the other purine. The binding ligand is further stabilized to the basal edge of the STING side chain by a direct and water-mediated hydrogen bond network. The phosphate backbone of the cGAMP ring system and the ribose hydroxyl group are stabilized by hydrogen bonds. The associated amino acids are located below the binding ligands (Gao et al., 2013c). This allows stably binding of cGAMP to the STING dimer.
As mentioned above, the binding of STING and cGAMP results in a butterfly-like dimer, in which the LBD of STING is 180° opposite its TM domain and exposes a CTT (Ergun et al., 2019; Shang et al., 2019). This conformation likely mediates the recruitment of coat protein complex II (COP-II), which, in turn, opens the “exit” for STING to leave the ER and transport through COP-II vesicles to the ER-Golgi intermediate compartment (ERGIC) and Golgi apparatus (Dobbs et al., 2015; Gui et al., 2019; Prabakaran et al., 2018). The importance of STING translocation in signaling implies that the TM domain of STING plays a role in regulating its activity (Shang et al., 2019).
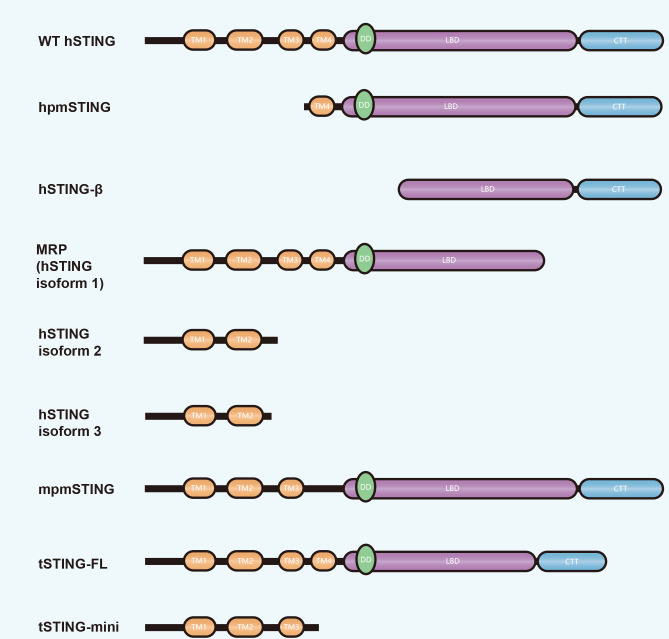
In addition to full-length STING that functions in cGAS-STING signaling, recent studies have also reported the existence of several STING isoforms in humans and other species due to selective splicing of pre-mRNA, although most are detrimental to STING-mediated immune responses due to incomplete functional domains (Liang et al., 2021) (Figure 2). To date, three human STING (hSTING) isoforms have been identified due to exon 7 deletion (i.e., hSTING isoform1, 2 and 3). Isoform1, also known as MITA-related protein (MRP), lacks part of the LBD for cGAMP binding as well as the entire CTT for TBK1 binding, while isoform2 and 3 contain only two TMs (TM1 and TM2) and cannot bind to TBK1 and activate downstream signaling. STING isoform transcription is preferred to full-length STING during HSV-1 infection, reducing the ratio of hSTING wild-type (WT)/isoforms and competitively inhibiting hSTING-WT-mediated IFN-I production (Chen et al., 2014; Rodríguez-García et al., 2018). However, MRP also retains some of its antiviral function. During hepatitis B virus (HBV) infection, MRP activates the NF-κB pathway and inhibits HBV replication (Liu et al., 2017b). A hSTING isoform lacking TM1, TM2, and TM3 is present in humans, known as human plasmatic membrane STING (hpmSTING), while that in mice (mpmSTING) lacks only one TM (TM4). Both hpmSTING and mpmSTING are restricted to the plasma membrane, with their CTT directed to the extracellular matrix, allowing them to sense extracellular cGAMP directly (Li et al., 2022c). In addition, a hSTING isoform lacking four TMs, called STING-β, is widely expressed in various human tissues and can be induced during viral infection to interact with WT STING and TBK1 to inhibit the induction of IFN-I (Wang et al., 2018b).
Figure 2.
Domain composition of STING isoforms in different species
Differences in domain composition between wild-type (WT) STING and STING isoforms are shown. TM: Transmembrane domain; DD: Dimerization domain; LBD: Ligand-binding domain; CTT: C-terminal tail.
A STING isoform is also found in Tupaia that benefits mediated innate immune responses, unlike the above-mentioned counterparts (Figure 2). Structurally, full-length Tupaia STING (tSTING-FL) is similar to that of hSTING, with four TMs and a CTT, and retains classical resistance to DNA viruses. The tSTING-FL isoform (tSTING-mini) retains only three TMs and can act upon RNA viral infection. During Newcastle disease virus (NDV) infection, tSTING-mini responds more rapidly than tSTING-FL and interacts with tIRF3 to promote phosphorylation and downstream IFN-I production (Xu et al., 2020a).
DOWNSTREAM SIGNALING
Upon activation by cGAMP, STING further interacts with other adaptor molecules and induces the phosphorylation and nuclear translocation of IRFs, leading to IFN production and antiviral or antidamage immune responses (Figure 3). In metazoans, activated STING mediates downstream signaling by recruiting and activating TBK1. STING binds to TBK1 through the CTT to form an interface, and mutations in STING Pro371Gln and TBK1 Gln581Ala at this interface then abrogate the interactions of STING with TBK1 and indirectly abrogate the phosphorylation and dimerization of IRF3 and the induction of downstream IFN-I (Zhang et al., 2019a). Based on analysis of the STING structure domain, IRF3 dimerization is induced by a STING fragment encompassing residues 281–379 (Tanaka & Chen, 2012). Multiple innate immunity adaptors, including mitochondrial antiviral signal protein (MAVS), Toll/interleukin 1 (IL-1) receptor domain containing inducing IFN-β (TRIF), and STING, have a highly conserved motif (p-Leu-X-Ile-Ser, where “p-” signifies phosphorylation and “X” suggests any amino acid) that is phosphorylated in response to ligand activation. Phosphorylation of this motif provides so-called “preparatory” phosphorylation of IRF3 by TBK1 (Chen et al., 2016c). Interestingly, a STING fragment containing only 39 residues (341–379) is sufficient to activate IRF3. Activity can be eliminated by deleting two amino acids from the C-terminal. Defective binding to IRF3 may be responsible for the inability of Ser366Ala and Leu374Ala mutant STING to promote IRF3 phosphorylation via TBK1 (Tanaka & Chen, 2012). Hence, conserved residues (i.e., Ser366 and Leu374) within STING are crucial for signal transduction (Christensen et al., 2016; Tanaka & Chen, 2012).
Figure 3.
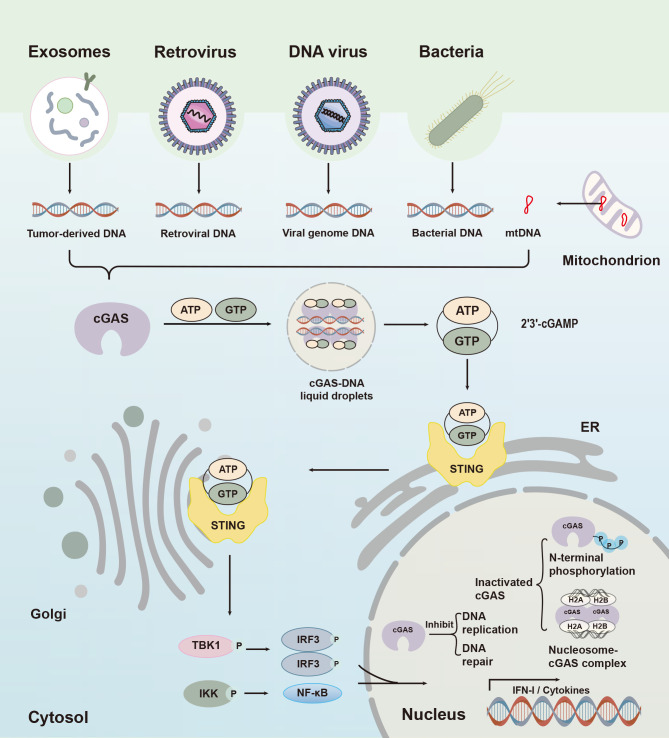
Cytoplasmic cGAS-STING signaling and nuclear cGAS function
Cytoplasmic cGAS recognizes foreign pathogens or damaged DNA to initiate relative immune responses. Upon binding to dsDNA, cGAS recruits ATP and GTP and catalyzes synthesis of 2′-3′cGAMP. This second messenger then binds to and activates ER-resident protein STING, which is then translocated from the ER to Golgi apparatus and induces recruitment of TBK1/IKK. TBK1 induces IFN-I production by promoting dimerization of IRF3 and IKK by mediating NF-κB entry into the nucleus. In the nucleus, cGAS is normally in an inhibited state due to chromatin tethering and hyperphosphorylation at the N-terminus, which functions to regulate DNA replication, inhibit DNA repair, and initiate nuclear innate immunity.
Induction of IFN-I through the cGAS-STING pathway also involves the activation of NF-κB (Figure 3). The key to activating NF-κB signaling is the inhibitor of κB kinase (IKK) complex, which consists of two catalytic subunits, i.e., IKKα and IKKβ (Abe & Barber, 2014). Specifically, IKKα and IKKβ phosphorylate inhibitory IκB, leading to its proteasome degradation, which, in turn, releases NF-κB transcription factor subunits such as p50 and p65 for nucleus translocation (Chen & Greene, 2004; Tu et al., 2013). NF-κB mediates the induction of IFN-I in the nucleus via positive regulatory domain II (PRD II) (Tu et al., 2013). After completing its role, STING is subsequently phosphorylated by serine/threonine UNC-51-like kinase (ULK1/ATG1) at position Ser366 (Konno et al., 2013) and translocated to the lysosome for degradation (Gonugunta et al., 2017). The translocation process is mediated by a class of vesicles called clathrin-coated vesicles (CCV), and adaptor protein complex-1 (AP-1) on the CCV (Liu et al., 2022). Therefore, activation of the cGAS-STING signaling pathway promotes activation and nuclear translocation of transcription factors such as NF-κB, IRF3, thereby stimulating the production of proinflammatory cytokines and IFN-I.
PHYSIOLOGICAL FUNCTIONS
Physiological functions of cytosolic cGAS-STING
Antipathogen immune responses: cGAS functions to recognize exogenous DNA and initiate antiviral immune responses. cGAS can bind to the DNA of a variety of viruses, including cytomegalovirus (CMV), Kaposi sarcoma-associated herpesvirus (KSHV), adenovirus, HSV, and cowpox virus (CPV) (Lio et al., 2016; Ma et al., 2015; Paijo et al., 2016; Rasaiyaah et al., 2013; Wu et al., 2015; Zhang et al., 2016a), and induces IFN production in a STING-dependent manner. For example, MS1 endothelial cells and RAW264.7 macrophages from cGAS, STING, and TBK1 knockdown mice show significant suppression of antiviral responses after adenovirus infection, suggesting that cGAS acts as a major DNA receptor that recognizes adenovirus infection (Lam et al., 2014). In addition, the cGAS-STING pathway also induces the production of small amounts of IFN in the resting state, which is important for the prevention and control of DNA and RNA virus infection in vivo (Chen et al., 2016c).
Although the cGAS-STING pathway is normally activated by DNA viruses, it may also initiate an immune response to RNA virus infection, potentially leading to “leakage” of cellular DNA and activation of the cGAS pathway against RNA virus infection. Cell membrane fusion can also trigger an innate immune response via STING-dependent DNA-independent sensing pathways (Foley, 2012). Induction of chemokine (C-X-C motif) ligand 10 (CXCL10) by virus-like particles (VLPs) or liposomes is abolished in both peritoneal cells (PC) and bone marrow-derived dendritic cells (BMDCs) of STING-deficient mice, which may contribute to immune defense against RNA viruses (Holm et al., 2012).
Retroviruses can also activate cGAS-STING innate immune signaling. The RNA of viruses or reverse transcription products is detected by RNA sensing receptors such as RIG-I via the cGAS-cGAMP-STING pathway, thereby triggering a second wave of continuous signaling and promoting the production of specific immunoglobulin M (Zeng et al., 2014). Human immunodeficiency virus (HIV) infection triggers the cGAS-STING signaling pathway, resulting in the production of IFN-I and other cytokines (Gao et al., 2013a). Notably, virus-induced IFN-I induction is eliminated by inhibition of HIV reverse transcriptase rather than integrase, demonstrating that reverse-transcribed HIV DNA stimulates the innate immune response. cGAS knockout (KO) mice or human cell lines prevent HIV, murine leukemia virus, and simian immunodeficiency virus from inducing cytokines, further supporting the hypothesis that cGAS is an inherent immunosensor of HIV and retrovirus (Gao et al., 2013a).
Cytosolic cGAS also participates as an immunological DNA sensor for bacteria, including Mycobacterium, Legionella, Listeria, and Shigella. Upon detection of cytoplasmic bacterial DNA, cGAS activates inherent antibacterial defenses of the host and promotes autophagy targeting bacteria such as Mycobacterium tuberculosis (Watson et al., 2015). Mice lacking cGAS are more susceptible to M. tuberculosis infection and exhibit higher mortality (Collins et al., 2015). In addition, Listeria DNA, but not cyclic di-AMP, stimulates IFN-I responses through a pathway dependent on cGAS-STING and DNA sensor IFI16 in macrophages (Hansen et al., 2014), and induction of IFN-I by most bacteria is eliminated in the absence of cGAS. STING alone can also induce the body to fight pathogens. Bacterially produced cyclic dinucleotides (CDNs), such as cyclic di-GMP and cyclic di-AMP, can directly activate STING and induce antibacterial responses (Chen et al., 2016c). Cyclic-di-AMP in live gram-positive bacteria can induce ER stress via STING, followed by inactivation of mammalian target of rapamycin protein (mTOR), leading to autophagy. Autophagy further alleviates ER stress and reduces phagocytic cell death, thereby maintaining cellular homeostasis (Moretti et al., 2017).
Recognition of damage-associated mitochondrial DNA (mtDNA): MtDNA leakage, cellular damage-associated stress observed in many physiologically processes, such as infection, disease, and aging, is also recognized by cGAS, which triggers activation of downstream damage-associated immune responses. When subjected to various stimuli, the amount of mtDNA changes and may leak from the mitochondria (De Gaetano et al., 2021; West et al., 2015). Currently, two possible mechanisms for mtDNA leakage have been demonstrated (Kim et al., 2019; McArthur et al., 2018). During apoptosis, BCL-2-associated X protein (BAX) and BCL-2 antagonist/killer (BAK) are activated and oligomerized on the outer mitochondrial membrane, leading to complete mitochondrial outer membrane permeabilization and subsequent release of mtDNA and apoptotic factors such as cytochrome C (McArthur et al., 2018). In addition, voltage-dependent anion channel (VDAC) proteins can also oligomerize on the outer mitochondrial membrane, leading to pore formation on the membrane (Kim et al., 2019). Mitochondrial membrane instability allows mtDNA to pass through and be exposed to the cytoplasm for recognition by cGAS (Kim et al., 2019; McArthur et al., 2018).
External stimuli, such as HSV-1 infection, can disrupt mtDNA homeostasis and induce faulty mtDNA packaging, leading to mtDNA leakage into the cytoplasm and activation of cGAS-STING signaling (West et al., 2015). In addition, pathogenic bacteria such as M. tuberculosis can activate cGAS and downstream production of IFN-β by mediating the release of mtDNA (Wiens & Ernst, 2016). Exogenous IL-1β is also capable of inducing mtDNA release, activating cGAS, and initiating downstream immune responses (Aarreberg et al., 2019). In an odontoblast inflammation model, lipopolysaccharide (LPS) causes mtDNA leakage and cGAS-STING activation, producing inflammatory factors such as IL-6 and inducing downstream inflammatory responses (Zhou et al., 2021a). Endogenous stimuli, such as intracellular pyrimidine imbalance, may also contribute to mtDNA leakage. Intracellular pyrimidine synthesis is a process dependent on the mitochondrial protease YME1-like ATPase (YME1L). MtDNA release is induced by the absence of intracellular pyrimidines due to YME1L deletion (Sprenger et al., 2021). In addition, mutations in encoding ATPase family AAA domain-containing protein 3A (ATAD3A) may also induce mtDNA release into the cytoplasm, thereby mediating cGAS-STING signaling pathway activation and interference stimulating gene (ISG) expression (Lepelley et al., 2021).
Cellular senescence: Cellular senescence refers to the gradual decline in cell proliferation, differentiation, and physiological functions with the passage of time in the process of performing life activities (Regulski, 2017). It acts as a natural tumor-prevention barrier and is also involved in fibrosis and tissue healing (Campisi, 2013). cGAS also plays a role in senescence (Loo et al., 2020).
DNA damage leads to the accumulation of cytoplasmic DNA, a hallmark event leading to senescence. Accumulation of DNA leads to the activation of cGAS and expression of senescence-associated secretory phenotypes (SASP) and inflammatory factors. Loss of cGAS accelerates spontaneous immortalization of mouse embryonic fibroblasts and abrogates SASP induced by DNA damage, suggesting that cGAS mediates cellular senescence and delays immortalization (Yang et al., 2017a). Recent research also identified a non-classical cGAS-STING pathway associated with cellular senescence. cGAMP binding to STING does not activate TBK1, but directly activates the ER-localized kinase STING-PKR-like endoplasmic reticulum kinase (PERK) (Zhang et al., 2022). PERK activation phosphorylates eukaryotic translation initiation factor 2 subunit α (eIF2α) and mediates cellular senescence and organ fibrosis (Zhang et al., 2022). Given the importance of the cGAS-STING pathway in cellular senescence, inhibitors of cGAS or STING may be very useful in the treatment of senility and age-related illnesses.
Physiological functions of nuclear cGAS
Nuclear-localized cGAS has distinct functions from cytoplasmic cGAS, and most are STING-independent (Figure 3).
DNA replication regulation: DNA replication is a rigorous and conserved process, abnormalities of which can lead to the development of various diseases. During DNA replication, the DNA double strand is separated by DNA helicases, after which various replication-associated proteins and enzymes bind to the unzipped sites to form a replication fork, which moves along the DNA to complete the replication process (Dewar & Walter, 2017). Nuclear cGAS binds to chromatin and regulates DNA replication in a DNA binding-dependent manner. The binding of cGAS reduces the movement rate of the replication fork, while deletion of cGAS accelerates movement but compromises DNA stability, leaving the cells under replication stress and highly sensitive to radiotherapy and chemotherapy. Therefore, targeted delivery of cGAS inhibitors to tumor lesions may be an effective antitumor treatment option (Chen et al., 2020).
DNA repair inhibition: DNA repair is one of the most important biological processes employed by cells to mitigate the deleterious effects of DNA damage. Research has shown that cGAS can prevent DNA replication by inhibiting homologous recombination (HR). Specifically, DNA double-strand breaks (DSBs) can induce importin α-dependent nuclear translocation of cGAS, in which cGAS is recruited to DSBs and interacts with PARP1 via poly(ADP-ribose). These cGAS-PARP1 interactions prevent the formation of PARP1-TIMELESS complexes, thereby inhibiting HR-mediated DNA repair and promoting tumor growth (Liu et al., 2018a). A critical step in the HR process is RAD51-dependent DNA strand invasion. cGAS can compress template DNA into a highly ordered state, thereby impeding DNA strand invasion and inhibiting HR-mediated DNA repair (Cui et al., 2020). Furthermore, cGAS can promote replicative cellular senescence by interacting with CDK1, blocking RNF8 recruitment, and inhibiting mitotic non-homologous end joining (NHEJ). Loss of cGAS may lead to end-to-end fusion between short telomeres and genomic instability (Li et al., 2022d). Cancer cells usually circumvent replicative senescence due to telomere damage by reactivating telomerase activity (Hwang, 2002), and promoting replicative senescence in cancer cells may be a potential new therapeutic direction. Whether this replicative function of cGAS can be exploited for cancer treatment deserves further investigation.
Innate immunity: Nuclear-localized cGAS can inhibit DNA sensing, DNA susceptibility, and enzymatic activity (Gentili et al., 2019; Michalski et al., 2020; Zhao et al., 2020), and is involved in innate immunity (Bai & Liu, 2022). DNA virus infection may result in the shedding and release of chromatin-bound cGAS into the nuclear soluble fraction. Nuclear soluble cGAS can sense viral DNA like cytoplasmic cGAS and mediate downstream IFN-I production, which, in turn, inhibits viral DNA replication (Wu et al., 2022). Nuclear cGAS in dendric cells (DCs) can produce cGAMP after infection with HSV-1, although cGAMP levels are 200-fold lower than cytoplasmic cGAMP levels produced by herring testis DNA (HT-DNA) transfection (Gentili et al., 2019). For the retrovirus HIV, viral perception by nuclear cGAS depends on the non-POU domain-containing octamer-binding protein (NONO). Nuclear cGAS interacts with NONO, which recognizes the nucleated HIV capsid and recruits HIV-DNA to the vicinity of nuclear cGAS, thereby enhancing cGAS perception of DNA and mediating innate immune response (Lahaye et al., 2018). Surprisingly, RNA viruses can also activate the innate immune response through nuclear cGAS, but not through reverse transcription of DNA by RNA viruses, nor is it dependent on the enzymatic activity of cGAS. Mechanistically, nuclear cGAS interacts with PRMT5 and facilitates the regulation of histone H3 arginine 2 by PRMT5. Symmetrical dimethylation of histone H3 arginine 2 at the IFN-I promoter promotes the entry of activated IRF3 into the IFN-I promoter, enhancing IFN-I and CXCL10 production and innate immune response (Cui et al., 2020). In the future, certain diseases caused by RNA viruses and retroviruses may find suitable treatment options through cGAS.
EVOLUTIONARY CONSERVATION OF CGAS AND STING
In addition to the detailed study of human and mouse cGAS-STING signaling, many cGAS and STING homologues have been identified and analyzed in other invertebrate and vertebrate species (De Falco et al., 2022; Liu et al., 2020; Oliveira et al., 2021; Wu et al., 2014; Yang et al., 2021). The origin of cGAS and STING can be traced to choanoflagellate Monosiga brevicollis (Wu et al., 2014). Within metazoans, cGAS and STING homologues are found in cnidarians, some arthropods, and nearly all chordates (except Takifugu rubripes), but are absent in nematodes and flatworms. Gene duplication events may have occurred for cGAS and STING, with two cGAS homologues found in Hydra magnipapillata (three for STING), Tribolium castaneum, several Drosophila species (e.g., D. virilis, D. persimilis, and D. pseudoobscura), Branchiostoma floridae (two for STING), and Danio rerio (Liu et al., 2020). During evolution, the functional domains of cGAS and STING have been primarily conserved, with slight differences. The length of the N-terminal domain tends to be shorter in invertebrate than vertebrate cGAS, and the NTase core and Mab21 domain are conserved among species, with the exception of M. brevicollis and invertebrate cGAS, which do not have a Zn-binding motif within the Mab21 domain. For residues critical for DNA binding and dimer formation, three residues essential for primary DNA binding (Lys384, Lys411, and Lys407 of hcGAS), three residues on the dimer surface (Lys347, Lys394, and Lys398 of hcGAS), and two residues on the second DNA-binding surface (Arg236 and Lys254 of hcGAS) are highly conserved in vertebrates but not in early-branching species (Gao et al., 2013b, 2013c).
For the STING domains, four TMs are conserved in M. brevicollis and metazoans, except for some arthropods, birds, and cephalochordate homologues, which have fewer than four TMs, and three arthropod homologues (Ixodes scapularis, Tribolium castaneum, and Apis mellifera), which lack TMs altogether (Fischer et al., 2020; Jiang et al., 2019, 2022; Zhang et al., 2016c). The LBD is conserved in all STING homologues and the CTT is present in all vertebrates, except for amphibian species Xenopus tropicalis and X. laevis. Structurally, STING homologues from other species are similar to those in mammals, and all are expected to have the ability to bind to cGAMP, although lower species may have weaker affinity. Among the six essential residues in the cGAMP binding pocket of STING (Tyr167, Arg238, Tyr240, Asn242, Glu260, and Thr263 of hcGAS), Tyr167 and Arg238 are fully conserved and Glu260 and Thr263 are highly conserved during evolution, while Tyr240 and Asn242 are only conserved in mammals (Gao et al., 2013c).
REGULATION OF CGAS-STING SIGNALING
Post-translational modification of cGAS and STING
cGAS-STING signaling is tightly regulated to prevent deleterious effects on the host from excessive responses. Aberrant IFN induction due to cytosolic cGAS activation is associated with autoimmune diseases (Decout et al., 2021). Therefore, tightly regulated initiation and progression of cGAS-STING signaling is crucial for eliciting an appropriate immune response. Positive regulatory mechanisms lead to rapid induction of IFNs and pro-inflammatory cytokines, whereas negative regulatory loops stand by to prevent excessive immune responses. Post-translational modifications represent an important cellular mechanism in regulating or fine-tuning cGAS-STING signal transduction (Figure 4) (Zhou et al., 2017).
Figure 4.
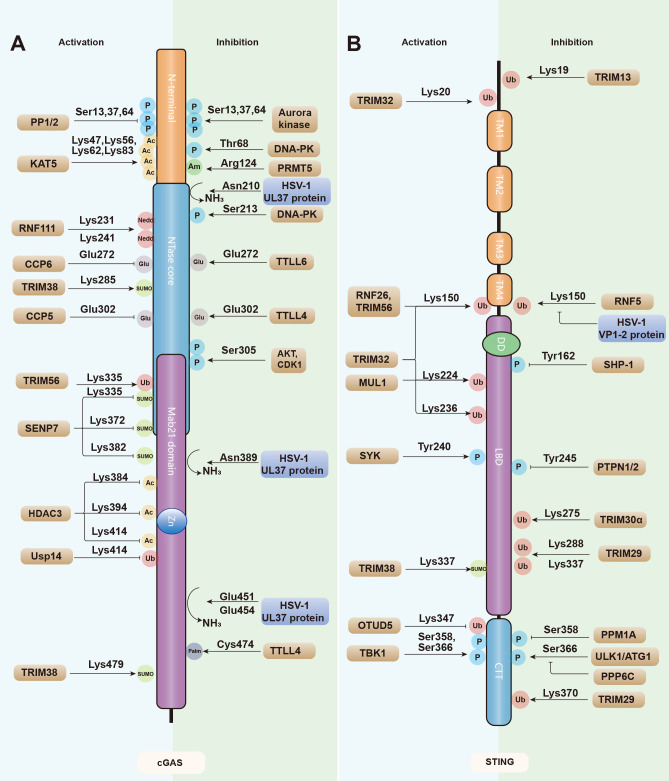
Post-translational modifications of cGAS and STING
A, B: cGAS (A) and STING (B) are regulated by a series of modifications, including activating/enhancing modifications (left) and inhibitory modifications (right). P, phosphorylation; Ub, ubiquitination; SUMO, SUMOylation; Glu, glutamylation; Ac, acetylation; Nedd, neddylation; Am, Arginine methylation; Palm, palmitoylation. Enzymes mediating cGAS-STING regulation are shown in brown and viral proteins are shown in blue. TM: transmembrane domain; DD: dimerization domain; LBD: ligand-binding domain; CTT: C-terminal tail.
Phosphorylation of cGAS: cGAS activity is well tuned to ensure timely and appropriate resolution of immune responses. Seo et al. (2015) showed that Akt kinases play a negative role in cGAS-mediated antiviral immune responses. Akt mediates the phosphorylation of Ser291/Ser305 of mcGAS/hcGAS, which is located in the enzymatic domain of cGAS in proximity to the catalytic site. This phosphorylation by Akt strongly inhibits cGAMP synthetization during HSV1 infection and activated Akt expression leads to a decrease in cGAMP and IFN-I production and an increase in HSV1 replication, while treatment with Akt inhibitors boosts cGAS-mediated antiviral immune signaling (Seo et al., 2015). DNA-dependent protein kinase (DNA-PK) specifically interacts with cGAS and phosphorylates cGAS at Thr68 to impair dimerization and at Ser213 to inhibit enzymatic activity during vesicular stomatitis virus (VSV) or HSV-1 infection (Sun et al., 2020).
How do cells avoid activation of cGAS by chromatin DNA exposure during mitosis? Two enzymes have been found to phosphorylate cGAS upon nuclear envelope breakdown (NEBD). A mitotic kinase CDK1-cyclinB complex phosphorylates the Ser305 site of cGAS, preventing activation of cGAS by host genomic DNA (Zhong et al., 2020). Aurora kinase B hyperphosphorylates the N-terminal Ser13 and Ser64 sites of cGAS. As the N-terminal of cGAS is an important domain that facilitates cGAS and DNA phase separation into droplets, its hyperphosphorylation directly prevents the formation of cGAS and DNA droplets, thereby affecting the oligomerization and activation of cGAS (Li et al., 2021b). Once mitosis is completed, protein phosphatases PP1 and PP2 may mediate the dephosphorylation of cGAS, restoring the ability of cGAS to sense DNA (Li et al., 2021b; Zhong et al., 2020).
Phosphorylation and dephosphorylation of STING: Phosphorylation of STING itself is necessary for downstream signal transduction, and STING contains two conserved serine and threonine sites, which are phosphorylation targets of TBK1. Structurally, the CTT of STING inserts into the TBK1 dimer groove, which is formed by the kinase domain of one TBK1 and dimer domain of the other. TBK1 phosphorylates STING at Ser 366, which, in turn, mediates the recruitment of IRF3 and activates the IFN-I pathway (Liu et al., 2015a; Zhang et al., 2019a). In addition, protein phosphatase 6 catalytic subunit (PPP6C) interacts and dephosphorylates STING at Ser366. The phosphorylation of STING is enhanced by the deletion of PPP6C, further inhibiting viral replication (Ni et al., 2020). TBK1 also phosphorylates STING at Ser 358 to promote its aggregation, while its dephosphorylation by protein phosphatase magnesium dependent 1A (PPM1A) antagonizes STING dimerization and inhibits its function (Li et al., 2015). In addition, tyrosine protein phosphatase non-receptor type (PTPN1/2) dephosphorylates STING at Tyr 245 and promotes proteasome degradation, thus inhibiting IFN-I induction (Xia et al., 2019).
STING phosphorylation also contributes to its proper translocation from the ER to the Golgi apparatus. Stimulation of cGAMP leads to STING and epidermal growth factor receptor (EGFR) interactions, followed by autophosphorylation as a prerequisite for the recruitment of tyrosine kinase Syk. Syk then phosphorylates STING at Tyr240, followed by STING translocation (Wang et al., 2020a, 2021b). STING transport is also regulated by myotubularin-related protein (MTMR) 3 and MTMR4, two protein tyrosine phosphatases that dephosphorylate the 3' position of phosphatidylinositol (PtdIns). Double knockdown of MTMR3 and MTMR4 results in the accumulation of PtdIns3P, which enhances the trafficking of ER-located STING to the Golgi apparatus, thereby facilitating antiviral responses (Dewi Pamungkas Putri et al., 2019).
Phosphorylation of STING not only contributes to the turning on of immune responses but is also involved in preventing STING overactivation and maintaining homeostasis. After delivering TBK1 to the endochromosomal/lysosomal compartment, STING is subsequently phosphorylated at Ser366 by ULK1/ATG1. Phosphorylation of STING at this time occurs in the autophagosome, promoting its degradation to prevent ongoing IFN-I production (Konno et al., 2013).
Glutamylation: Glutamylation is an ATP-dependent process that adds glutamate chains to conserved glutamate residues in target proteins (Janke et al., 2008). The addition of glutamate is regulated by tubulin tyrosine ligase (TTL) and tubulin tyrosine ligase-like (TTLL) enzymes. Glutamate can further be removed by cytoplasmic carboxypeptidases (CCPs), and thus glutamylation is also a reversible post-translational modification (Ablasser, 2016; Garnham et al., 2015; Rogowski et al., 2010). Xia et al. (2016) screened the TTL and CCP families for enzymes involved in regulating cGAS glutamylation and found that TTLL4 mono-glutamylated cGAS at Glu302 and TTLL6 poly-glutamylated cGAS at Glu272 can lead to the blockade of cGAMP synthesis and loss of cGAS-DNA binding capacity, respectively. Glutamylation of cGAS ultimately inhibits the production of IFN-I and host immune response to viral infection. The CCP family member CCP5 further mediates the removal of the glutamate chain from Glu302 and CCP6 for the hydrolysis of the glutamate chain of Glu272, thereby restoring cGAS activity and enhancing IRF3 activation and IFN-I production (Xia et al., 2016).
Acetylation and deacetylation: Acetylation can also control cGAS activity (Dai et al., 2019; Song et al., 2020). Lysine acetyltransferase 5 (KAT5) is capable of acetylating cGAS at several sites, including Lys47, Lys56, Lys62, and Lys83 in the N-terminal structural domain, and promotes cGAS-DNA binding activity and activation (Song et al., 2020). In contrast, acetylation at other sites may help maintain cGAS in the inactive state, thereby preventing cGAMP synthetization and autoimmune diseases. For example, aspirin, a non-steroidal anti-inflammatory drug that promotes protein acetylation (Vane & Botting, 2003), can directly acetylate at the Lys384 and/or Lys394 and Lys414 sites, which are located at the domain of DNA binding, catalytic activity, and dimerization. Acetylation of cGAS at these sites prevents its activation and subsequent immune response. Histone deacetylase 3 (HDAC3) is involved in the deacetylation of Lys384 and promotes the activation of cGAS to some extent. However, deacetylases for the Lys394 and Lys414 acetylated sites remain to be discovered (Dai et al., 2019).
Ubiquitination and deubiquitination of cGAS: Ubiquitin is a small, highly conserved protein containing 76 amino acids, with seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and one methionine residue (M1) (Park et al., 2014). Ubiquitination is a powerful post-translational modification that is usually achieved by E1 activating enzymes, E2 conjugating enzymes, and E3 protein ligases that mediate the binding of the ubiquitin chain to target proteins (Deng et al., 2020a). Mechanistically, ubiquitin first forms a thioester bond with E1 and then transfers to E2. The E2 enzyme-ubiquitin complex then interacts with the E3 ubiquitin ligase to form an isopeptide bond between the carboxyl group of Gly-76 at the terminus of ubiquitin and the ε-amino group of the lysine residue in the substrate (Malynn & Ma, 2010; Song & Luo, 2019; Varshavsky, 2017). As a versatile post-translational modification, ubiquitination plays a pivotal role in regulating the cGAS-STING signaling pathway, which is essential for the tight regulation of mediated antiviral responses.
The first E3 ubiquitin ligase identified for cGAS ubiquitination was ring finger protein 185 (RNF185), which contains a RING domain that interacts with cGAS. RNF185 specifically catalyzes K27-linked ubiquitination and promotes cGAS enzymatic activity (Wang et al., 2017c). Deletion of RNF185 attenuates cytoplasmic DNA-induced activation of IRF3 (Wang et al., 2017c). The RING finger protein interacting with C kinase (RINCK), another well-known E3 ubiquitin ligase, activates cGAS by mediating its monoubiquitination. The presence of RINCK is required for cGAMP synthesis, and therefore is also important for the production of downstream IFN-I. Cells lacking RINCK are significantly compromised in defense against DNA viruses (Liu et al., 2018c). TNF receptor associated factor 6 (TRAF6), another E3 ubiquitin ligase, also specifically interacts with cGAS and mediates its ubiquitination, thereby promoting cGAS activation and downstream IFN-I production (Chen & Chen, 2019). TRIM56, a member of the tripartite motif (TRIM) E3 ubiquitin ligase family, also actively regulates cGAS. The C-terminal NCL1-HT2A-Lin41 (NHL) region of TRIM56 regulates interactions with the N-terminal of cGAS, mediates the ubiquitination of cGAS at Lys335, and enhances cGAS dimerization, DNA binding, and cGAMP production, thereby promoting antiviral immune responses (Seo et al., 2018).
As ubiquitination is a reversible process and added ubiquitins can be removed by deubiquitinating enzymes (DUBs), deubiquitination is essential for the functional regulation of cGAS (Snyder & Silva, 2021). Several DUBs regulate the stability and function of cGAS (Chen et al., 2016a; Guo et al., 2019), with ubiquitin-specific peptidase 27X (USP27X) the first DUB identified to interact with cGAS and mediate its deubiquitination. Zhang et al. (2020b) found that the ubiquitin carboxyl-terminal hydrolase (UCH) structural domain of USP29 and C-terminal structure of the cGAS NTase domain mediate USP29-cGAS interactions, and USP29 interacts with cGAS in a sustained manner. USP27X and USP29 have a similar mechanism for the deubiquitination of cGAS, both preventing the degradation of cGAS via the proteasomal pathway by removing the K48-linked ubiquitin chain of cGAS and maintaining cGAS protein levels. Hence, USP27X and USP29 promote the production of cGAMP and downstream IFNs to mediate inflammation and autoimmune responses (Guo et al., 2019; Zhang et al., 2020b).
Alternatively, some DUBs do not regulate cGAS deubiquitination through direct interactions (Chen et al., 2016a). For example, when the organism is infected by HSV-1, the recruitment of USP14 leads to TRIM14 cleavage of the k48-linked ubiquitin chain of cGAS located at the Lys414 site, thereby inhibiting p62-mediated autophagic degradation of cGAS, maintaining the stability of cGAS, regulating IFN-I expression, and enhancing antiviral capacity (Chen et al., 2016a).
Ubiquitination and deubiquitination of STING: STING undergoes K6, K11, K27, K48, and K63-linked polyubiquitination to fine-tune its function during immune response (Deng et al., 2020a). The E3 ubiquitin ligase TRIM13 possesses a TM domain containing an ER locus that binds directly to STING and catalyzes the polyubiquitination of K6-linked STING at the Lys19 site. This polyubiquitination facilitates the degradation of STING and maintains hemostasis in the event of abnormal STING activation (Li et al., 2022e). RNF26 interacts with STING through its RING domain and promotes K11-linked polyubiquitination at the Lys150 site. Deletion of RNF26 inhibits IFN-I production downstream of STING and weakens the host antiviral immune response (Qin et al., 2014). In addition, the ER protein autocrine motility factor receptor (AMFR) is recruited in an insulin-inducible gene 1 (INSIG1)-dependent manner and interacts with STING in response to cytoplasmic DNA stimulation (Wang et al., 2014). AMFR and INSIG1 form an E3 ubiquitin ligase complex, which catalyzes K27-linked polyubiquitination. This modification serves as an anchoring platform for TBK1 recruitment and as a translocation platform to facilitate its transfer to extranuclear microsomes. Depletion of AMFR or INSIG1 impairs STING-mediated induction of antiviral genes, an important and previously unexplored link in the STING signaling pathway (Wang et al., 2014).
Several E3 ligases also mediate K48-linked ubiquitination of STING, namely TRIM29, TRIM30α, RNF5, and RNF90, all of which act as negative feedback regulators of STING, but with slightly different mechanisms and modification sites (Li et al., 2018a; Wang et al., 2015; Yang et al., 2020; Zhong et al., 2009). TRIM29 mediates ubiquitination at Lys288, Lys337, and Lys370 by interacting with the CTT of STING, while TRIM30α mediates ubiquitination at Lys275, leading to proteasome-dependent STING degradation (Li et al., 2018a; Wang et al., 2015; Xing et al., 2017). RNF5 interacts with and ubiquitinates STING at the Lys150 site in a virus infection-dependent manner, promoting its degradation and regulating its homeostasis in conjunction with RNF26 (Qin et al., 2014). RNF90 interacts with STING through its RING structural domain and recognizes and degrades STING through the proteasome system (Yang et al., 2020).
K63-linked ubiquitination is also essential for the functional regulation of STING (Akira et al., 2012). Several enzymes mediate STING K63-linked ubiquitination, including TRIM32, TRIM56, and mitochondrial E3 ubiquitin ligase (MUL) 1 (Ni et al., 2017; Tsuchida et al., 2010; Zhang et al., 2012). TRIM32-mediated K63-linked ubiquitination of STING at the Lys20/150/224/236 site significantly enhances STING-induced IFN-I production by facilitating its interaction with TBK1 in the mitochondria and ER (Zhang et al., 2012). In addition, TRIM56 interacts with ubiquitin regulatory X domain-containing protein (UBXN) and STING, thereby promoting K63 ubiquitination of STING at Lys150. This modification induces STING dimerization, a prerequisite for recruitment of antiviral kinase TBK1 (Tsuchida et al., 2010; Yang et al., 2018). MUL1 ubiquitinates STING at Lys224, and further mediates the translocation and phosphorylation of STING, activation of downstream TBK1 and IRF3, and subsequent host antiviral response (Ni et al., 2017). Different kinds of ubiquitination may coordinate with each other to properly mount antipathogen immune response mediated by STING (Malynn & Ma, 2010). For example, death associated protein kinase 3 (DAPK3), an antitumor immune kinase, balances the ubiquitination state of STING under different physiological conditions. In the resting state, DAPK3 inhibits polyubiquitination and promotes the degradation of STING K48 linkages to ensure timely inactivation and activation of STING to mediate downstream IFN-I production upon external stimuli. After viral invasion, upstream cGAS synthesizes the second messenger cGAMP, stimulates DAPK3 to phosphorylate the E3 ligase LMO7 on Ser863, and further mediates K63-linked ubiquitination of STING, thereby mediating STING dimerization and downstream antiviral response (Takahashi et al., 2021).
Deubiquitination of STING is mediated by various DUBs, most of which belong to the ubiquitin-specific protease (USP) and ovarian tumor protease (OTU) families (Li et al., 2019). The USP family of DUBs is very important, with USP13, USP18, USP20, USP21, USP44, and USP49 shown to mediate the deubiquitination of STING (Chen et al., 2021). Among them, USP13 and USP21 inhibit TBK1 recruitment and IFN-I induction by removing the STING K27-linked polyubiquitin chain (Chen et al., 2017; Sun et al., 2017). USP18, USP20, and USP44 mediate hydrolysis of the STING K48-linked ubiquitin chain. USP18 does not interact directly with STING but maintains normal phosphorylation of downstream IRF3 and induction of IFN-I by recruiting USP20 to catalyze the K33/K48 linkage ubiquitination of STING and to protect STING from proteasome-dependent degradation (Zhang et al., 2016b, 2019b). In contrast, USP44 interacts with STING directly upon DNA virus infection to remove the K48-linked polyubiquitin chain at the Lys236 site. Mice lacking USP44 exhibit weakened STING expression and reduced IFN-I and proinflammatory factor induction, and are more susceptible to HSV infection, thus demonstrating the importance of USP44 in maintaining STING stability (Zhang et al., 2020a). In addition, after DNA virus infection, USP21 and USP49 inhibit STING oligomerization and recruitment of downstream TBK1 by removing the STING K63-linked ubiquitin chain, thus inhibiting IFN-I induction and antiviral response (Chen et al., 2017; Ye et al., 2019).
DUB eukaryotic translation initiation factor 3 subunit 5 (EIF3S5) and ovarian tumor domain-containing protein 5 (OTUD5) can remove the K48-linked polyubiquitin chain of STING. EIF3S5 is recruited by inactive rhomboid protein 2 (iRhom2) and interacts with STING (Luo et al., 2016). OTUD5 can cleave the K48-linked polyubiquitin chain at the STING Lys347 site. Both enzymes protect STING from degradation and are extremely important for its stability and downstream innate immune responses (Guo et al., 2021). Myb-like, SWIRM, and MPN domains 1 (MYSM1), an enzyme earlier identified as an H2A deubiquitinating complex (Zhu et al., 2007), interacts with STING and inhibits its activity by removing K63-linked ubiquitination. Deletion of MYSM1 results in normal expression of IFN-I and proinflammatory factors (Tian et al., 2020).
These ubiquitinating and deubiquitinating enzymes are critical for regulating the cGAS-STING pathway and understanding their mechanisms of action may facilitate the study of therapeutic options for autoimmune diseases.
SUMOylation and deSUMOylation: Similar to ubiquitin, small ubiquitin-like modifier (SUMO) proteins form covalent bonds with the branched chain of lysine of target proteins, a process known as SUMOylation. SUMOylation can affect protein interactions, alter protein subcellular localization, and enhance or diminish protein stability (Yang et al., 2017b). SUMOylation is also involved in the regulation of many biological processes, including DNA damage repair, cellular immunity, apoptosis, and carcinogenesis (Eifler & Vertegaal, 2015; Flotho & Melchior, 2013; Han et al., 2018; Rabellino et al., 2017; Wang & Dasso, 2009). In recent years, the regulatory mechanisms of SUMOylation on the cGAS-STING signaling pathway have been investigated.
cGAS is SUMOylated at multiple sites, with differences in the spatiotemporal regulation of cGAS activity by different ligase-mediated SUMOylation. The ubiquitin ligase TRIM38 targets cGAS for SUMOylation in an enzymatic activity-dependent manner in uninfected cells and the early stages of viral infection (Hu et al., 2016). SUMOylation of cGAS inhibits its polyubiquitination and degradation. cGAS is also SUMOylated by TRIM38 at Lys285 (Lys271 in mcGAS) and Lys479 (Lys464 in mcGAS), and SUMOylation at Lys464 stabilizes mcGAS activity (Hu et al., 2016). In addition, sentrin/SUMO-specific protease 7 (SENP7) interacts with cGAS and promotes its activation, mediated by the deSUMOylation of SENP7 (Cui et al., 2017). Mechanistically, SUMO couples to residues Lys335, 372, and 382 located in the DNA-binding sites of cGAS, and the binding of SUMO to these residues inhibits the binding of cGAS to DNA, thereby impacting cGAS function. Knockdown of SENP7 reverses this situation by catalyzing the deSUMOylation of cGAS, supported by the finding that knockdown of SENP7 predisposes mice to HSV-1 infection (Cui et al., 2017).
In terms of STING regulation, TRIM38 also promotes STING activation and protein stability during the early stages of viral infection (Hu et al., 2016). SUMOylation of STING at Lys337 can mask its 326QEVLR330 motif, which is essential for chaperone-mediated autophagy (CMA). Thus, SUMOylation of STING can inhibit its own degradation and facilitate its recruitment to IRF3. In contrast, at the late stages of infection, cGAS and STING are deSUMOylated by Senp2 and subsequently degraded via the proteasomal and CMA autophagic pathways, respectively, avoiding the organismal stress response caused by the continuous activation of cGAS and STING (Hu et al., 2016).
Neddylation: In addition to ubiquitination and SUMOylation, neural precursor cell expressed, developmentally downregulated 8 (NEDD8) also regulates the function cGAS and STING (Enchev et al., 2015). NEDD8 is a ubiquitin like protein (UBL) and completes neddylation by covalently coupling its carboxy-terminal glycine to the lysine residues of target proteins via specific NEDD8 E1 (heterodimer of APPBP1 and UBA3), E2 (well-characterized Ubc12 and less characterized UBE2F) (Rabut & Peter, 2008), and shared ubiquitination E3 ligases (Enchev et al., 2015).
RNF111 of the NEDD8 E3 ligase family is essential for cGAS activation (Li et al., 2021a). RNF111 neddylates cGAS at the Lys231 and Lys241 sites, thereby enhancing its ability to bind DNA and promoting formation of the cGAS-DNA 2:2 complex. Inhibition of NEDD8 E2 enzymatic activity with the neddylation inhibitor MLN4924 significantly impairs IFN-I and proinflammatory cytokine induction. Thus, neddylation plays a crucial role in the regulation of the cGAS-STING pathway and the antiviral function of cells. However, as a reversible process, the NEDD8 isopeptidases responsible for deneddylation of cGAS remain to be characterized (Li et al., 2021a).
Arginine methylation: Arginine methylation modification is the covalent labeling of proteins by the addition of methyl groups to arginine residues (Blanc & Richard, 2017). Modification plays an important role in the regulation of protein function and DNA binding (Bedford & Clarke, 2009). PRTM5, a member of the protein arginine methyltransferase (PRMT) family, is an “initiator” of arginine methylation modification of cGAS (Ma et al., 2021). PRMT5 can directly interact with cGAS through its RGG/RG motif and transfer a methyl group from S-adenosylmethionine to Arg124 of cGAS. Subsequent symmetric dimethylation of the Arg124 residue blocks the binding of cGAS to DNA, severely inhibits the enzymatic activity of cGAS, and ultimately leads to a weakened antiviral immune response (Ma et al., 2021).
Palmitoylation: Protein palmitoylation is a powerful and conserved post-translational modification in which fatty acid chains are attached to cysteine residues of proteins via reversible thioester bonds (Wang et al., 2020c). This post-translational modification plays a crucial role in regulating protein stability, cell membrane trafficking, and enzymatic activity (Liu et al., 2017a). The interactions of proteins with lipids and other proteins in the membrane cavity are influenced by the presence of palmitate on the protein (Chen et al., 2016c; Linder & Deschenes, 2007). Protein palmitoylation is catalyzed by DHHC-palmitoyltransferase, and palmitoylation of cGAS can negatively regulate its function. Mechanistically, Zn finger DHHC domain-containing protein 18 (ZDHHC18) palmitoylates cGAS at Cys474, thereby reducing the interaction between cGAS and dsDNA and inhibiting the dimerization of cGAS (Shi et al., 2022). While the powerful functions of palmitoylation are well documented (Zhou et al., 2017), reports on the palmitoylation of cGAS remain scarce, which may be a hot topic for future research.
A recent study reported that the ER-related protein ZDHHC1 is a positive regulator of virus-triggered STING-dependent immunological responses (Zhou et al., 2014). STING dimerization and downstream TBK1 and IRF3 signaling are mediated by ZDHHC1, which is connected to the composition of STING (Zhou et al., 2014). Palmitoylation of STING at the Golgi apparatus is required for its self-activation. The palmitoylation inhibitor 2-bromopalmitate (2-BP) prevents STING from being palmitoylated and thus hinders the mediated IFN-I response. STING is palmitoylated at two cystine sites, Cys88 and Cys 91, located at the proximal end of the membrane, and STING palmitoylation is inhibited when these two residues are mutated (Broz & Dixit, 2016). Cys91 palmitoylation is easier than that of Cys88 as the latter is buried within TM2–TM3 of STING, and palmitoylation of TM3-proximitized Cys91 may promote tetramer formation, aiding STING activation and subsequent immune response (Zhang et al., 2019a).
Pathogen escape by kidnapping post-translational modifications of cGAS-STING: As post-translational modification plays a critical role in regulating the cGAS-STING signaling pathway, viruses have developed their own strategy to hijack this modification to avoid clearance by cGAS-STING-launched immune responses (Zhou et al., 2017). The UL37 protein of HSV-1 deamidates mcGAS at Asn196, Asn377, Gln436, and Gln439 (Asn210, Asn389, Gln451, and Gln454 in hcGAS), which are all located in the Mab21 enzyme domain of cGAS. Deamidation of cGAS by UL37 inhibits the cGAMP synthesis but does not affect dsDNA-binding, dimerization, or GTP-ATP-binding activity, and therefore inhibits antiviral responses of the host and allows HSV-1 to replicate and proliferate (Zhang et al., 2018). In addition, the HIV-1 nonstructural protein viral infectivity factor (Vif) can recruit the cellular tyrosine phosphatase SHP-1 to STING and mediate STING dephosphorylation at Tyr162. Tyr162 dephosphorylation further inhibits cGAS K63-linked Lys337 ubiquitination, resulting in the failure of STING oligomerization and downstream IRF3 activation, thereby allowing HIV to evade host immunity (Wang et al., 2022b).
Human T lymphotropic virus type 1 (HTLV-1), HBV, HSV, porcine circovirus 2 (PCV2), and other viruses can hijack STING ubiquitination to escape host immune defenses (Bodda et al., 2020; Chen et al., 2017; Liu et al., 2015c; Wang et al., 2017b; Wu et al., 2021b). The viral polymerase (Pol) of HBV inhibits STING K63-linked polyubiquitination through its reverse transcriptase (RT) and RNase H (RH) structural domains (Liu et al., 2015c). The Tax protein of HTLV-1 interacts with the CTT of STING to block STING K63-linked ubiquitination (Wang et al., 2017b). The VP1-2 protein of HSV-1 exerts deubiquitinase activity to remove the K63-linked ubiquitin chain from STING Lys150 made by TRIM32 (Bodda et al., 2020). In addition, during late HSV-1 infection, the virus hijacks USP21 to hydrolyze the STING K27/63-linked ubiquitin chain, thereby inhibiting IFN-I production (Chen et al., 2017). Viruses can also inhibit the cGAS-STING signaling pathway by affecting the phosphorylation of STING. For example, the viral IFN regulatory factor 1 (vIRF1) protein of KSHV can inhibit downstream IFN production by inhibiting the interaction of STING with TBK1 and subsequent phosphorylation (Ma et al., 2015). KSHV can also evade the immune response by manipulating PPM1G. The tegument protein open reading frame 33 (ORF33) of KSHV dephosphorylates STING by enhancing the recruitment of PPM1G, ultimately inhibiting the antiviral response of the host (Yu et al., 2020b).
Correct subcellular localization and transport
As an ER-resident protein, correct localization and proper trafficking of STING are critical for the integrity of the signaling pathway (Ishikawa & Barber, 2008; Shang et al., 2019) and are mediated by the Ca2+ sensor stromal interaction molecule 1 (STIM1) regulator. STIM1 retains STING on the ER membrane, and co-expression of full-length STIM1 or STING-interacting fragments of STIM1 suppresses the function of dominant STING mutants that cause autoinflammatory diseases. STIM1 deficiency can lead to spontaneous activation and enhanced expression of IFN-I in mice and patients with combined immunodeficiency (Srikanth et al., 2019). In addition, loss of STIM1 strongly enhances IFN-I expression after viral infection and prevents mortality caused by DNA virus infection in vivo (Srikanth et al., 2019).
The Shigella effector protein IpaJ efficiently blocks STING signal translocation from the ER to ERGIC, a rate-limiting event in STING signal transduction (Dobbs et al., 2015). In addition, STING mutations associated with human autoimmunity (such as Asn153Ser, Val147Leu, Asn154Ser, Val155Met, and Arg232Ala) can lead to constitutive ER withdrawal and activation of STING unrelated to cGAMP binding (Dobbs et al., 2015). Sar1 is a small GTPase that controls the transport of COP-II from the ER to the Golgi apparatus. Sar1 knockdown can prevent STING translocation from the ER and TBK1 binding to STING (Ogawa et al., 2018). After exposure to viral infection, excessive production of cGAMP may inhibit STING transport. cGAMP mediates the condensation of STING into a membranous biocondensate, and the degree of STING condensation is positively correlated with the concentration of cGAMP (Yu et al., 2021). Condensed STING is localized to the ER and forms a puzzle structure in which STING can recruit TBK1, but neither STING nor TBK1 can be phosphorylated or mediate the activation of IRF3 (Yu et al., 2021). These studies suggest that correct transport of STING is important for signaling pathway regulation.
Apoptosis and autophagy
The role of the cGAS-STING signaling pathway in host innate immunity is self-evident, however its overreaction can also cause immune diseases (Gao et al., 2015), e.g., lupus erythematosus. Therefore, important mechanisms such as apoptosis and autophagy are needed to maintain cGAS-STING signaling pathway stability and prevent its overactivation (Galluzzi & Green, 2019).
Apoptosis is defined as regulated and orderly cell death to preserve internal environment stability (D’Arcy, 2019). When the cell undergoes apoptosis, the nuclear membrane ruptures and nuclear DNA is released, which can activate cGAS and induce IFN-I production. Active caspase cascades that promote cell apoptosis also prevent dead cells from activating the host immune response (White et al., 2014). Activated caspase-3 cleaves cGAS to prevent excessive cytokine generation during virus-induced apoptosis and DNA leakage (Ning et al., 2019). In detail, caspase-3 cleaves Asp319, a conserved site of hcGAS positioned on the central β sheet in the catalytic pocket of cGAS, to inactivate enzymatic activity, thereby protecting cells from autoimmune responses induced by released nuclear DNA during apoptosis (Wu & Li, 2020).
Autophagy can be divided into three main types, i.e., microautophagy, CMA, and macroautophagy. Macroautophagy is a conserved stress response in which components to be phagocytosed are encapsulated in a bimodal vesicle for degradation (Galluzzi & Green, 2019). The autophagy involved in cGAS-STING is mainly macroautophagy.
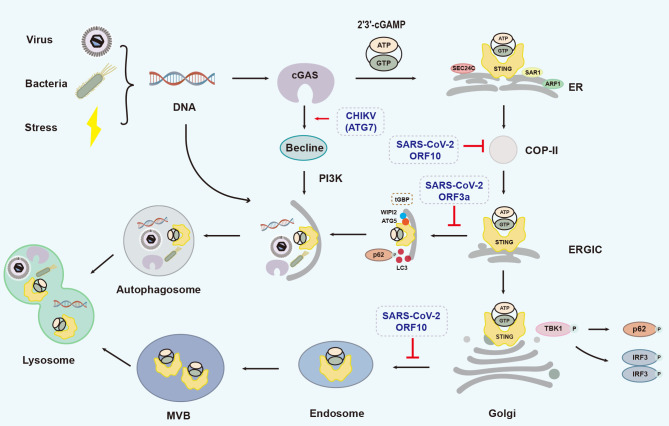
When dsDNA activates the cGAS-STING signaling pathway, the autophagy-mediated resolution strategies for cGAS and STING are different. cGAS directly interacts with the autophagy protein Beclin-1, followed by increased activity of phosphatidylinositol 3-kinase class III (PI3K) and formation of autophagosomes, hence limiting excessive cGAS activation and severe inflammatory reactions (Liang et al., 2014). STING is also involved in TBK1-dependent and -independent autophagy. Upon DNA invasion, cGAS-synthesized cGAMP induces STING activation and LC3 lipidation. In detail, cGAMP recruits secretion-associated Ras-related 1 (SAR1), SEC24 homolog C, COPII coat complex component (SEC24C), and ADP ribosylation factor 1 (ARF1) to form the COP-II complex (Ge et al., 2013, 2014), which assists in the transfer of STING from ER to ERGIC. STING-containing ERGIC further acts as a membrane source for LC3 lipidation and induces LC3 lipidation dependent on WD-repeat domain PtdIns(3)P-interacting2 (WIPI2) and ATG5 (Brandizzi & Barlowe, 2013; Yang et al., 2019). Lipidated LC3-positive membranes wrap around STING and pathogenic DNA to form an autophagosome, independent of TBK1 (Gui et al., 2019). In addition, ERGIC-located STING can also be transported to the Golgi apparatus, where it activates TBK1 and IRF3. At this time, P62 is phosphorylated by TBK1 to mediate the transport of Golgi-located STING to the autophagy-associated vesicular endosome, which further forms a multi-vesicular body (MVB). Finally, MVBs and autophagosomes integrate with lysosomes and degrade the contents in a Ras-related protein 7 (Rab7)-dependent manner (Gui et al., 2019; Prabakaran et al., 2018) (Figure 5).
Figure 5.
Autophagic processes involving cGAS-STING, adapted from Gui et al. (2019) with some modifications
Pathogen- or damage-derived dsDNA activates the cGAS-STING signaling pathway, cGAS directly interacts with Beclin-1 and increases PI3K activity to induce autophagy and avoid excessive cGAS activation. cGAMP synthesized by cGAS induces STING translocation from ER to ERGIC in a SAR1, SEC24C, and ARF1-dependent manner. STING-containing ERGIC further acts as a membrane source for LC3 lipidation and induces LC3 lipidation relying on WIPI2 and ATG5. In addition, ERGIC-located STING can also transport to the Golgi apparatus, and P62 mediates its further transportation to autophagy-associated vesicular endosome and lysosomes for degradation in a MVB and small GTPase Rab7-dependent manner. MVB: multi-vesicular body. Gray dashed boxes indicate viral proteins that promote (→) or inhibit (┸) processes. Brown dashed box shows that Tupaia guanylate-binding protein (tGBP) promotes autophagy together with WIPI2 and ATG5.
cGAS-STING-associated autophagy can also be regulated by viral infection (Figure 5). For example, upon exposure to the RNA Chikungunya virus (CHIKV), the capsid protein of CHIKV mediates the degradation of cGAS through ATG7-dependent autophagy (Yang et al., 2019). Furthermore, ORF3a and ORF10 of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can bind to STING and interrupt the autophagic process (Han et al., 2022; Su et al., 2022). ORF3a can disrupt the interaction between STING and LC3 and ORF10 can inhibit the transport of STING from the ER to ERGIC, thereby blocking STING-mediated autophagy, affecting pathogenic DNA degradation, and antagonizing the antiviral response (Han et al., 2022; Su et al., 2022). In the presence of HSV-1 infection, mSTING can mediate the antiviral response through IFN-I-independent autophagy (Yamashiro et al., 2020). On the other hand, tSTING promotes autophagy in response to HSV-1 by binding to Tupaia guanylate-binding proteins (tGBPs) and cooperating with Tupaia p62 and Tupaia LC3 (Gu et al., 2021). Upon exposure to Zika virus (ZIKV), Drosophila STING (dSTING) can induce Drosophila autophagy in the brain to inhibit ZIKV replication (Liu et al., 2018b).
CGAS-STING SIGNALING IN DISEASE
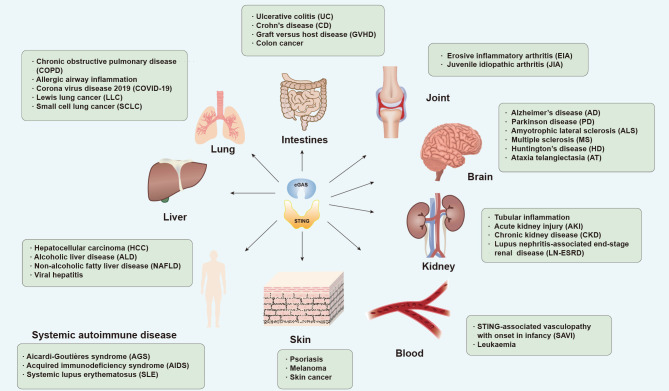
Although the function of the cGAS-STING pathway in initiating innate immunity was unraveled first, recent studies have also explored its role in the pathogenesis of disease (Figure 6).
Figure 6.
cGAS-STING and related diseases
Dysregulation of cGAS-STING pathway is associated with various diseases. Representative diseases in different organs are summarized.
Autoimmune diseases
Chronic inflammation and autoimmune diseases, including Aicardi-Goutières syndrome (AGS), juvenile idiopathic arthritis (JIA), and systemic lupus erythematosus (SLE), can develop when the inflammatory immune response goes unresolved and persists for a long period of time (Okin & Medzhitov, 2012). As early as the 1980s, biologists discovered that excessive IFN-I in mice may have adverse effects on the host (Gresser et al., 1980). Up-regulation of IFN-I by the cGAS-STING signaling pathway is central to the pathogenesis of autoimmune syndromes (Crow, 2015). At the genetic level, these syndromes are associated with genetic mutations in several key genes, including adenosine deaminases acting on RNA (ADAR1), IFN induced by helicase C domain 1 (IFIH1), three prime repair exonuclease 1 (TREX1), SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1), and ribonuclease H2 subunit A (RNaseH2). TREX1 is a 3' repair exonuclease that degrades cytoplasmic DNA. Mutations in the TREX1 gene can cause AGS (Gray et al., 2015). TREX1-/- and RNaseH2-/- mice exhibit autoimmune and inflammatory phenotypes after cGAS-STING pathway activation, which is related to increased ISG expression (Gray et al., 2015). Furthermore, loss of cGAS in Trex1-/- mice results in complete lethality, with significantly reduced tissue inflammation and autoantibody production failure, indicating that cGAS may be a key factor in autoimmune disease (Gray et al., 2015). Gain-of-function mutations in STING can cause autoinflammatory disease, i.e., STING-associated vasculopathy with onset in infancy (SAVI), and transcription of IFN-I in peripheral blood mononuclear cells of SAVI patients is elevated, accompanied by skin inflammation, lung damage, and breathing difficulties (Liu et al., 2014). In addition, the lysosome-localized DNaseII enzyme removes DNA from dead cells and DNA expelled from the nucleus. Absence of this enzyme can lead to the accumulation of DNA and increased activation of STING signaling and IFN-I production, leading to autoinflammation (Baum et al., 2017). In DNaseII/IFN-α/β/ω receptor (IFNαR) double KO mice, the long bones and spleen are two sites of erythropoiesis and local DNA accumulation. Osteocyte recruitment and induction of inflammatory factors, key factors in erosive inflammatory arthritis (EIA), are also enhanced at these two sites. In contrast, knockdown of STING significantly inhibits these responses and rescues the systemic autoinflammatory response (Baum et al., 2017).
cGAS-STING has also been implicated in autoimmune hepatitis (Apel et al., 2021). Notably, in mice treated with lectin concanavalin A, a model of autoimmune hepatitis, neutrophil extracellular traps (NETs) activate cGAS in cytoplasmic lysates and induce IFN-I production. Activation of this pathway is mediated by NET-associated serine protease neutrophil elastase (NE) (Apel et al., 2021). However, the detailed mechanisms of the role of cGAS-STING in autoimmune hepatitis remain to be elucidated.
Inflammatory bowel disease (IBD)
The intestinal microbiota maintains intestinal homeostasis, and maturation of the mucosal immune system is accompanied by microbial colonization. Disruption of intestinal immune system homeostasis can lead to IBD, including ulcerative colitis (UC) and Crohn’s disease (CD) (Baumgart & Sandborn, 2012; Ordás et al., 2012). STING is critical for maintaining intestinal homeostasis. In mice, STING deletion causes decreased mucus formation, decreased antibody release, and changes in the gut microbial composition (Canesso et al., 2018). The STING pathway is involved in maintaining intestinal barrier integrity and reducing the severity of graft-versus-host disease (GVHD) (Fischer et al., 2017). Furthermore, STING in mononuclear phagocytes, primarily controlled by commensal bacteria, can trigger production of the anti-inflammatory cytokine IL-10, and reduce intestinal inflammation (Ahn et al., 2017).
However, excessive activation of STING is counterproductive. Treatment of mice with the STING agonist DMXAA significantly worsens dextrose sulfate (DSS)-induced colonic injury and induces differentiation of bone marrow-derived macrophages (BMDM) toward proinflammatory subtypes (Martin et al., 2019). Furthermore, ER stress and the resulting autophagy can cause intestinal epithelial dysfunction, intestinal ecological dysregulation, and proinflammatory response (Benjamin et al., 2013; Larabi et al., 2020). For example, deletion of X-box-binding protein 1 (XBP1) in intestinal epithelial cells (IECs) can induce ER stress and colitis (Kaser et al., 2008). Deletion of autophagy-related 16-like 1 (ATG16L1), an IBD risk gene capable of regulating the IL-22 signaling pathway, exacerbates ER stress and subsequent STING-dependent IFN-I induction, leading to necrotizing epithelial cell death (Aden et al., 2018). Autophagy and ER stress can exacerbate activation of the cGAS-STING-IFN pathway and more severe inflammatory responses. Thus, genes associated with ER stress are promising for studies on therapeutic regimens targeting cGAS-STING.
Inflammatory lung disease
Leakage of self-DNA and dysregulation of the cGAS-STING pathway may lead to lung inflammation and fibrosis. For example, silica induces the release of self-DNA following lung cell death, activates the cGAS-STING signaling pathway, and induces downstream proinflammatory IFN-I expression, leading to pneumonia (Benmerzoug et al., 2018). Cigarette smoke causes the release of auto-DNA following damage of the respiratory barrier and activation of cGAS-STING signaling, ultimately leading to chronic obstructive pulmonary disease (COPD) (Nascimento et al., 2019). cGAS may also promote inflammation by regulating granulocyte-macrophage colony-stimulating factor (GM-CSF) production in airway epithelial cells. Knockdown of cGAS significantly attenuates ovalbumin (OVA) or house dust mite (HDM)-induced allergic airway inflammation (Han et al., 2020). These findings suggest that the cGAS-STING pathway is a driver of inflammatory lung disease.
SARS-CoV-2 is the causative agent of the COVID-19 global epidemic (Berthelot & Lioté, 2020; Driggin et al., 2020). SARS-CoV-2 infection and host cell angiotensin-converting enzyme 2 (ACE2) expression expose cytoplasmic chromatin during cell fusion, activating the cGAS-STING signaling pathway and contributing to antiviral responses. The STING agonist diABZI is effective in reducing viral load at the epithelial cell tip and reducing epithelial cell damage (Zhou et al., 2021b; Zhu et al., 2021). Novel STING agonist CDGSF can immunize against the SARS-CoV-2 spike protein, causing high titers of antibodies and enhancing antiviral responses (Wu et al., 2021a). However, cytoplasmic damage and self-DNA leakage caused by SARS-CoV-2 in epithelial cells may also result in excessive cGAS-STING activation, leading to excessive release of IFN-β and tissue factors that induce focal death of epithelial cells (Berthelot et al., 2020a, 2020b; Berthelot & Lioté, 2020; Deng et al., 2020b; Mdkhana et al., 2021). Severity of COVID-19 symptoms is related to age and underlying disease (e.g., diabetes), which may be partly due to abnormalities in cGAS-STING signaling with aging and metabolic disturbances (Berthelot & Lioté, 2020). In addition, the viral ORF3a co-protein of SARS-CoV-2 specifically binds to STING and blocks nuclear accumulation of p65 to inhibit NF-κB signaling and facilitate viral escape (Rui et al., 2021). The major 3CL protease of SARS-CoV-2 inhibits Lys63-linked ubiquitination of STING, therefore inhibiting the recruitment of TBK1 and blocking IFN-I production (Rui et al., 2021). ORF9b interacts with STING and TBK1 to block IRF3 phosphorylation and promote viral replication (Han et al., 2021). These findings increase our understanding of the pathogenesis of SARS-CoV-2, with important implications for new therapeutic strategies.
Hepatitis
Viral hepatitis: Viral hepatitis includes liver lesions caused by hepatitis A virus (HAV), HBV, HCV, HDV, HEV, and other non-hepatophilic viruses, which can further be classified as acute, chronic, severe, and biliary hepatitis based on manifestations (Castaneda et al., 2021). According to the World Health Organization (WHO), hepatitis B and C cause the highest mortality rate among hepatitis types (WHO, 2017). Hence, we focused on the role of the cGAS-STING signaling pathway in regulating hepatitis B and C below.
As a DNA virus, the HBV genome can be directly recognized by cGAS to induce ISG56 expression via the cGAS-STING signaling pathway, thus initiating the innate immune response, and inhibiting the assembly and replication of HBV (Dansako et al., 2016; He et al., 2016). However, failure of the host antiviral immune response is an important factor in hepatitis B progression. In human hepatoma (HepG2) cells and immortalized mouse hepatocytes, STING expression is extremely low and barely initiates any immune response (Guo et al., 2017; Karimi-Googheri et al., 2015). Antiviral signaling can be largely induced by the injection of STING agonist DMXAA (Guo et al., 2015) or by overexpression of STING (Lauterbach-Rivière et al., 2020), leading to proper cGAS-STING activation and IL6 production, as well as reduction in cytoplasmic viral nucleocapsids and inhibition of HBV replication (Dansako et al., 2019). HBV itself has also derived multiple ways to evade clearance by cGAS-STING signaling, e.g., encapsulating its genomic DNA (Verrier et al., 2018) and disrupting STING K63-linked polyubiquitination by its Pol protein (Liu et al., 2015c). The nonstructural protein 4B (NS4B) protein can directly bind to STING and use its N-terminal structure to disrupt the interaction between STING and TBK1, blocking TBK1 activation and inhibiting IFN-I production (Ding et al., 2013; Nitta et al., 2013; Yi et al., 2016). In addition, NS4B can also inhibit STING accumulation in a dose-dependent manner by reducing the sensitivity of STING to cGAMP through its TM domain (Yi et al., 2016).
Alcohol-related hepatitis: Alcoholic hepatitis is a form of alcoholic liver disease (ALD), the pathogenesis of which is centered on oxidative stress associated with ethanol metabolism and endotoxin production in Kupffer cells (Gao & Bataller, 2011). Early studies found that ethanol induces ER stress, activates STING, and phosphorylates IRF3, associated with the expression of pro-apoptotic molecule BAX and apoptosis of hepatocytes (Petrasek et al., 2013). Later experiments confirmed that cGAS-produced cGAMP is capable of intercellular transmission via hepatic gap junction protein (Cx32) and induces activation of IRF3 in neighboring cells, further amplifying alcohol-induced oxidative stress, inflammation, and hepatocyte injury (Luther et al., 2020).
Non-alcoholic fatty liver disease (NAFLD): Simple steatosis, non-alcoholic steatohepatitis (NASH), and cirrhosis are all classified as NAFLD (Xu et al., 2021). Up-regulation of STING and IRF expression can be detected in methionine-choline-deficient diet (MCD) and high-fat diet (HFD)-induced obese mice and free fatty acid (FFA)-induced L-O2 cell lines. Knockdown of STING and treatment with NF-κB inhibitor BAY11-7082 significantly reduce FFA-induced liver inflammation and apoptosis, as demonstrated by the detection of NF-κB signaling pathway-related molecules, inflammatory cytokines, and apoptosis signaling pathway-related molecules (Qiao et al., 2018; Yu et al., 2019). An increase in STING+ cells is reported in the livers of NASH patients, including monocyte-derived macrophages (CCR2+, S100A9+), blast cells (CD68+), and CD163+ macrophages (Wang et al., 2020b). Mechanistically, STING significantly increases the proinflammatory state of macrophages, and tumor necrosis factor α (TNF-α) and IL-6 expression levels are increased in mouse blastocytes, further increasing hepatocyte fat accumulation and inflammatory response (Luo et al., 2018; Yu et al., 2019). Furthermore, in a lean NAFLD mouse model, sterol regulatory element-binding protein cleavage activating protein (SCAP, a cholesterol sensor) can specifically recruit STING and TBK1 to the Golgi apparatus, inducing activation of the NF-κB pathway, and thus increasing inflammatory metastasis and fat accumulation (Huang et al., 2022). Iron is also able to activate the cGAS-STING signaling pathway to induce inflammation. Expression levels of cGAS, STING, and downstream IRF3 are significantly elevated in the livers of iron-treated mice, a novel mechanism of NASH (Li et al., 2022a). Based on the pathogenic mechanisms mentioned above, related drugs have been investigated and applied. For example, traditional Chinese medicine Ling Gui Zhu Gan can significantly improve HFD-induced hepatic steatosis by inhibiting the STING-TBK1 pathway in hepatic macrophages (Cao et al., 2022).
Nephritis
Chronic kidney disease (CKD) and acute kidney disease (AKD) pose a considerable threat to global public health (Levey et al., 2020). As a highly metabolically active organ rich in mitochondria, the typical pathology of the kidney is increased oxygen consumption and decreased electrolyte transport efficiency due to oxidative stress, leading to renal hypoxia, mitochondrial stress, and renal tubular inflammation (Bai & Liu, 2021). Studies of kidney cells from mitochondrial transcription factor A (TFAM) KO mice have revealed that aberrant packaging of mtDNA results in its translocation across the cell membrane, activation of the cGAS-STING pathway, and expression of inflammatory factors. In contrast, ablation of STING ameliorates renal fibrosis in a mouse model of CKD, indicating that the cGAS-STING pathway may play a role in CKD (Maekawa et al., 2019). Lupus nephritis (LN)-associated end-stage renal disease (LN-ESRD) is an immune-complex nephritis caused by SLE involvement of the kidney and is characterized by an increase in IFN-I and nucleosome-associated dsDNA (nsDNA) levels. cGAS recognizes nsDNA and activates IRF3 through STING, triggering the expression of apolipoprotein L1 (APOL1) and IFNβ. APOL1 expression induces renal disease, and therefore, inhibition of the cGAS-STING pathway may be an effective therapeutic option (Davis et al., 2019).
In an acute kidney injury (AKI) model, cisplatin treatment induces mtDNA leakage into the cell membrane through the BAX pore of the outer mitochondrial membrane in renal tubules, thereby activating the cGAS-STING pathway and triggering the inflammatory response that causes AKI (Maekawa et al., 2019). Treating cells with STING covalent antagonist H151 results in a significant reduction in the expression of proteins related to apoptosis and injury of renal tubular cells, as well as a significant improvement in cisplatin-induced mitochondrial damage and a decrease in mtDNA content. Therefore, drugs designed to inhibit STING-induced inflammatory response may be a potential treatment for AKI (Gong et al., 2021).
Neurodegenerative diseases
Neurodegenerative diseases are a group of central nervous system (CNS) disorders caused by damaged neurons and spinal cord sheaths, represented by progressive cognitive and behavioural dysfunction, and associated with oxidative stress, apoptosis, and neuroinflammation (Dugger & Dickson, 2017). Microglia are the dominant central intrinsic immune cells in the CNS. The function and state of microglia are closely linked to inflammation in the brain, while astrocytes respond to inflammatory signals to promote inflammation and participate in the regulation of multiple life processes of the nervous system under physiological and pathological states (Paul et al., 2021).
Activation of microglia by viruses or damaged mtDNA initiates the cGAS-STING signaling pathway, which secretes IFN-I and acts on neuronal IFN A receptors (IFNARs) to trigger neuronal antiviral defense mechanisms. However, overactivation of this pathway can lead to neuroinflammation (Cox et al., 2015; Linnerbauer et al., 2020) and common neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), multiple sclerosis (MS), Huntington’s disease (HD), and ataxia telangiectasia (AT).
AD is a neurodegenerative disease characterized by progressive cognitive decline and memory loss (Hansen et al., 2018b). Nicotinamide adenine dinucleotide (NAD+) is key to several processes in AD neurons, including DNA repair and mitochondrial autophagy, and plays a central role in cellular metabolism. The brains of AD mice exhibit low levels of NAD+ and elevated expression of cGAS and STING. When treated with NAD+ precursor nicotinamide riboside (NR), DNA released during mitophagy can be reduced, thereby inhibiting cGAS-STING activation, and reducing neuroinflammation and cellular senescence (Hou et al., 2021; Larrick & Mendelsohn, 2021). Thus, modulation of the cGAS-STING pathway may contribute to the treatment of AD patients.
PD is an autosomal recessive neurodegenerative disorder (Kitada et al., 1998). Mutations in the PTEN-induced kinase 1 (PINK1) or parkin gene may lead to familial PD (Valente et al., 2004). PINK1 is a ubiquitin kinase and parkin is an E3 ubiquitin ligase, both of which degrade damaged mitochondria in a mitochondrial autophagy-dependent manner. When PINK1 and parkin are absent, the damaged mitochondrial autophagic pathway is interrupted and leaked mtDNA accumulates and activates the cGAS-STING pathway, causing neuroinflammation and further induction of PD. This response can be alleviated by STING KO (Sliter et al., 2018). STING levels are higher in the substantia nigra of PD patients, supporting the conclusion that the cGAS-STING signaling pathway may be involved in the pathogenesis of PD (Jauhari et al., 2020).
Accumulation of TAR DNA-binding protein 43 (TDP-43) is an important hallmark of ALS, a neurodegenerative disease of the motor neurons (Al-Chalabi & Hardiman, 2013). Mechanistically, when excess TDP-43 accumulates in the cytoplasm, it enters the mitochondria and induces mtDNA release, further activating the cGAS-STING signaling pathway. Increased levels of downstream IFN-I further mediate the neuroinflammatory response. This response can be rescued by administration of cGAS or STING inhibitors or by knockdown of cGAS and STING (Lee & Woodruff, 2021; Yu et al., 2020a).
MS is a chronic inflammatory neurodegenerative disease characterized by demyelination (Reich et al., 2018; Song et al., 2019). The function of the cGAS-STING signaling pathway in mediating inflammatory responses makes it a potentially effective therapeutic option for further study. Particle-encapsulated cGAMP can enhance STING activation and promote IFN-I and IL-10 production in peripheral mononuclear blood cells of MS patients (Johnson et al., 2021). The ganciclovir (GCV) drug induces IFN-I responses in microglia via activation of the cGAS-STING signaling pathway, thereby inhibiting inflammation, and providing protection in MS (Mathur et al., 2017).
HD is a polyglutamine disorder caused by CAG trinucleotide repeats (genetic modifiers of HD (GeM-HD) Consortium, 2019). The brain region most affected by HD is the striatum, and pathology includes DNA damage, DNA repair, and mitochondrial dysfunction (Kim et al., 2021; Maiuri et al., 2021). Leaked mtDNA following mitochondrial dysfunction activates the cGAS-STING pathway, followed by a stronger autophagic response in HD striatal cells and an exacerbated inflammatory response in HD mice (Jauhari et al., 2020; Sharma et al., 2020). Toll-interacting protein (TOLLIP) directly interacts with STING and is important for regulating STING homeostasis. PolyQ-rich proteins in striatal cells of HD patients compete with TOLLIP, affecting the binding of TOLLIP to STING, leading to STING degradation. However, it is unclear whether degradation of STING contributes to HD remission, which will be an interesting area of research in the future (Pokatayev et al., 2020).
AT is a neurodegenerative disease with multiple inflammatory manifestations caused by ataxia-telangiectasia mutated kinase (ATM), an important DNA damage repair kinase, the loss of which inhibits DNA repair and leads to DNA accumulation, thereby inducing activation of the cGAS-STING signaling pathway and production of IFN-I and inflammatory factors (Cree et al., 2021; Reich et al., 2018). Analysis of brain microglia from AT mice suggests that loss of inflammasomes involving STING and melanoma 2 sensors leads to secretion of neurotoxic cytokines and neurotoxicity, and STING-specific inhibitor CCCP blocks activity and reduces proinflammatory cytokine secretion, alleviating disease (Härtlova et al., 2015).
Although abnormal activation of cGAS-STING signaling can lead to neurological diseases under certain pathological conditions, Wang et al. (2022a) also showed that cGAS-STING is involved in promoting nerve regeneration in response to nerve injury. Mechanistically, injured peripheral nervous system (PNS) axons secrete IFNγ, which up-regulates cGAS expression in Schwann and blood cells in a non-cell-autonomous manner. Subsequently, the immunotransmitter cGAMP produced by cGAS is sensed by neuronal STING and further mediates axonal regeneration. In addition, cGAS-produced cGAMP also has the potential to increase the expression of IFNγ and ISG in nerves, which not only inhibits viral transmission but may be beneficial for axonal regeneration in the CNS (Wang et al., 2022a). Therefore, proper activation and use of cGAS-STING signaling may also benefit the treatment of neuroinflammatory diseases.
Tumors
cGAS-STING in tumor prevention: cGAS-STING mediates the body’s immune response, and DNA from dead tumor cells is considered an important signal that triggers IFN induction (Corrales & Gajewski, 2016). Genomic instability, mutation or deletion of tumor suppressor genes, oxidative stress, and exuberant metabolism are often found in tumor cells, greatly increasing the possibility of DNA leakage into the cytoplasm and activation of the cGAS-STING pathway (Deng et al., 2014). In addition, DCs are the most important antigen-presenting cells in the tumor microenvironment, and activation of the cGAS-STING signaling pathway in the DC cytoplasm can promote cell maturation and mobilize tumor-specific T cells to promote antitumor immunity (Klarquist et al., 2014; Nagata et al., 2021). CD47 binds to SIRP-α on the surface of macrophages and specifically blocks antibody-mediated phagocytosis of tumor cells by macrophages, an important mechanism for tumor immune escape. By blocking the interaction of SIRPα with CD47, cGAS in DCs allows these cells to recognize tumor mitochondrial DNA faster than macrophages, thereby contributing to IFN-I production and antitumor adaptive immunity (Liu et al., 2015b; Xu et al., 2017). In addition, STING is essential for radiation-induced adaptive immune responses, and mice lacking STING show functional deficits in initiating spontaneous CD8+ T cells against tumors (Liu et al., 2015b; Woo et al., 2014). Exogenous IFN-I treatment can rescue cGAS deficiency and STING initiation. Activation of STING by the second messenger cGAMP can enhance radiation-induced antitumor immunity (Deng et al., 2014). In mouse models of melanoma and colon cancer, intratumoral injection of cGAMP forces activation of STING in DCs, significantly enhances antitumor CD8+ T responses, and controls contralateral tumor growth (Wang et al., 2017a). On the other hand, cGAMP can trigger neutrophil migration. Within tumors, activation of STING enhances neutrophil migration to the tumor in an NF-κB/chemokine (C-X-C motility site) ligand (CXCL1/2)-dependent manner. Blockade of neutrophil migration with anti-CXC chemokine receptor 2 (CXCR2) monoclonal antibodies further compromises the efficacy of intratumoral cGAMP treatment. These findings suggest that migration of neutrophils and induction of neutrophil IFN-I by cGAMP treatment are critical for T cell activation and antitumor effects in tumor-draining lymph nodes (dLNs). In vitro analyses have also shown that IFN-I enhances neutrophil cytotoxicity (Nagata et al., 2021). Activation of exogenous STING triggers an antitumor immune response by recruiting and activating neutrophils in tumors through the CXCL1/2 and IFN-I signaling pathways (Nagata et al., 2021). Furthermore, in small cell lung cancer (SCLC), targeting DNA damage response (DDR) proteins, such as poly ADP-ribose polymerase (PARP) and checkpoint kinase 1 (CHK1), significantly increases the expression of programmed death 1 ligand (PD-L1) (Sen et al., 2019). In addition, in several SCLC cell lines, DDR targeting induces high expression of chemokine CXCL10, mediated by activation of the cGAS-STING signaling pathway, which also leads to recruitment of CD8+ T cells and antitumor immunity (Sen et al., 2019). The combination of cGAMP and PD-L1 antibodies is also reported to have a potent antitumor effect (Wang et al., 2017a), as the ability of cGAMP to trigger antitumor immunity is further enhanced by blocking PD1 and cytotoxic T lymphocyte-associated protein 4 (CTLA4) (Demaria et al., 2015).
cGAS-STING in tumorigenesis: Contrary to popular understanding, cGAS-STING may also negatively regulate the antitumor immune response due to the inextricable relationship between DNA damage and cancer (Li & Chen, 2018). DNA damage in tumor cells mediates the translocation of nuclear cGAS and inhibits DNA repair, thereby promoting tumor growth (Liu et al., 2018a). In a mouse skin cancer model, the DNA damaging agent 7,12-dimethylbenz(a)anthracene (DMBA) can activate the cGAS-STING signaling pathway to induce skin carcinogenesis, while deletion of STING inhibits skin cancer growth (Ahn et al., 2014). Furthermore, DNA is sensed by STING in mouse lymphoid tissues, which induces IFN-I production, stimulates indoleamine-2,3-dioxygenase (IDOase) activity in DCs, activates regulatory T cells (Tregs), and suppresses T cell responses (Huang et al., 2013). Treatment with c-di-GMP enhances STING activation, which further induces selective expression of IFN-β in DCs and suppresses T helper type 1 (Th1) immune responses, thereby promoting tumor growth (Huang et al., 2013). STING-induced IDO activity in the tumor microenvironment also enhances Lewis lung cancer (LLC) growth. Notably, the absence of STING increases CD8+ T cell infiltration and tumor cell killing in LLC models, suggesting that STING signaling plays a role in attenuating the effects of CD8+ T cells during carcinogenesis (Lemos et al., 2016). In breast and lung cancer models, cancer cells with brain metastases are able to transport cGAMP to astrocytes in the brain and activate downstream signaling pathways to produce inflammatory factors, thereby promoting tumor growth (Chen et al., 2016b).
Chromosomal instability (CIN) can also promote the evolution and metastasis of tumor cells and can significantly hinder tumor therapy. Mechanistically, CIN can lead to the rupture of micronuclei and release of DNA into the cytoplasm, activating the cGAS-STING signaling pathway and inducing production of inflammatory factor IL-6, further activating signal transducer and activator of transcription 3 (STAT3) and promoting tumor cell growth (Hong et al., 2022).
As many cGAS-STING signaling pathways are closely related to tumors, relevant treatments are gradually being explored. Recent studies have shown that oxidized mtDNA and STING signals are important for increasing antitumor efficacy of radiation tumor cell vaccinations. The STING-IFN pathway is activated in DCs by oxidized mtDNA, which subsequently cross-presents irradiated tumor cell-derived antigens to CD8+ T cells, thus inducing antitumor immunity (Fang et al., 2021). In mouse cancer cells, ATM deletion activates the cGAS-STING pathway through down-regulation of mitochondrial TFAM, enhancing lymphocyte infiltration into the tumor microenvironment, leading to mtDNA leakage and cGAS-STING pathway enhancement. This may be an effective way to improve cancer immunotherapy (Hu et al., 2021). CDNs are novel immunostimulants that elicit robust CD8+ T cell-mediated antitumor responses in tumor models. Notably, CDNs can enhance both natural killer (NK) cell activation and cytotoxicity via the cGAS-STING-IFN response and indirectly enhance antitumor effects via IL-15 and its receptors (Nicolai et al., 2020). In clinical practice, glycoside antigen-based vaccines have been constructed using the non-nucleotide small molecule STING agonist diABZI, which not only enhances the production of antibodies and T cell immune responses, but also inhibits tumor growth in mice (Wang et al., 2021c). A novel nano-vaccine based on STING has also been developed, which fuses antigens to PC7ANP nanoparticles to stimulate cytotoxic T cell responses. Importantly, PC7ANP can facilitate the presentation of tumor antigens to antigen-presenting cells relying on STING, further activating the IFN-I response, and inducing antitumor immunity (Luo et al., 2017). However, STING agonists may induce resistance due to the expansion of immunosuppressive B cells blocking NK cells. For example, if cGAMP is used as an agonist, STING will induce B cells to express IL-35 in an IRF3-dependent manner, thus inhibiting the proliferation of NK cells and weakening the antitumor response. How to solve this challenge remains to be investigated (Li et al., 2022b). In other aspects, DNA damage induced by radiotherapy can result in cytoplasmic leakage, which may initiate the antitumor immune response in the host but is insufficient to activate cGAS-STING. Under this limitation, studies have demonstrated that synchronizing Mn2+ supply with accumulated cytoplasmic DNA following radiotherapy can boost cGAS-STING pathway activation, thereby improving RT-induced antitumor immunity (Wang et al., 2021a). Furthermore, as mentioned above, TREX1 mutations can cause AGS. This enzyme is highly expressed after high-dose radiotherapy to remove cytoplasmic DNA, while multiple low-dose radiotherapy reduces the induction of TREX1, accumulates cytoplasmic DNA, and promotes cGAS-STING pathway activation and antitumor immunity (Vanpouille-Box et al., 2017).
Other diseases
Abnormal activation of cGAS-STING signaling by HFD may contribute to the development of obesity-related diseases (Gong et al., 2020). Given the enrichment of saturated fatty acids such as palmitic acid, HFD can cause mitochondrial damage and DNA leakage into the cytoplasm. Leaked DNA stimulates the STING-IRF3 pathway via cGAS and increases the expression of intercellular adhesion molecule 1 (ICAM-1). The adipose-IRF3 pathway is activated in adipose tissue of HFD-fed obese mice (Mao et al., 2017). Knockdown of STING prevents adipose tissue inflammation, obesity, and glucose intolerance partly caused by overeating (Mao et al., 2017). Recent research has also shown that psoriasis, a chronic inflammatory skin disease, is associated with the STING pathway. Notably, various cytokines are associated with psoriasis, including IFN-γ, TNF-α, IL-17, and IL-22, which induce the up-regulation of IFN-inducible protein (IFI16) in keratin-forming cells through activation of STAT3 signaling. In turn, IFI16 can recruit STING and induce IFN-I production by detecting cytoplasmic DNA (Cao et al., 2016). Knockdown of mouse p204 (homologous to human IFI16) can inhibit epidermal proliferation in mice with psoriatic dermatitis induced by immunodissociative hormones (Cao et al., 2016).
THERAPEUTIC DRUGS TARGETING CGAS AND STING
As cGAS-STING signaling is relevant to the occurrence of a variety of diseases, small molecule drugs targeting the cGAS-STING signaling pathway are promising therapeutic candidates. Several drugs have been shown to modulate cGAS-STING and their well-defined regulatory mechanisms fall into several broad categories, including cGAS inhibitors and STING inhibitors. Inhibitors of cGAS (An et al., 2015, 2017, 2018; Chu et al., 2021; Lu et al., 2022; Steinhagen et al., 2018; Wang et al., 2018a) act primarily through three mechanisms, i.e., blocking cGAS-DNA binding, inhibiting cGAS catalytic sites (Hall et al., 2017; Lama et al., 2019; Tan et al., 2021; Vincent et al., 2017; Xu et al., 2020b), and affecting post-translational modifications of cGAS (Dai et al., 2019). Inhibitors of STING act by inhibiting STING-cGAMP binding (Hong et al., 2021; Li et al., 2018b; Siu et al., 2019) and affecting STING palmitoylation (Haag et al., 2018; Hansen et al., 2018a; Vinogradova et al., 2020), dimerization, or transportation (Gao et al., 2022). However, there are several drugs that promote STING activation and act as STING agonists, some of which are CDN analogues (Cavlar et al., 2013), while others activate STING by stabilizing a “closed” conformation similar to STING-cGAMP (Chin et al., 2020; Gao et al., 2013c; Pan et al., 2020; Ramanjulu et al., 2018; Song et al., 2021; Xi et al., 2020) or induce the formation of higher-order oligomers of STING (Lu et al., 2022; Pryde et al., 2021) (Table 1).
Table 1. Structures and targets of cGAS and STING drugs.
| Compound | Structure | Target | Reference |
| Inhibitors of cGAS | |||
| Quinacrine and related AMDs |
|
cGAS-DNA binding | An et al., 2015, 2017 |
| PAH |
|
cGAS-DNA binding | Chu et al., 2021 |
| X6 |
|
cGAS-DNA binding | An et al., 2018 |
| Suramin |
|
cGAS-DNA binding (human specific) | Wang et al., 2018a |
| Ru.521 |
|
cGAS active site (mouse specific) | Vincent et al., 2017; Xu et al., 2020b |
| PF-06928215 |
|
cGAS active site | Hall et al., 2017 |
| G150 |
|
cGAS active site (human specific) | Lama et al., 2019 |
| Compound 25 |
|
cGAS active site | Tan et al., 2021 |
| Aspirin |
|
Acetylation of cGAS | Dai et al., 2019 |
| Inhibitors of STING | |||
| Compound 18 |
|
STING-CDN binding pocket | Siu et al., 2019 |
| Astin C |
|
STING-CDN binding pocket (mouse specific) | Li et al., 2018b |
| SN-011 |
|
STING-CDN binding pocket | Hong et al., 2021 |
| NO2-FAs |
|
Palmitoylation of STING | Hansen et al., 2018a |
| C-176 |
|
Palmitoylation of STING (mouse specific) | Haag et al., 2018 |
| C-178 |
|
Palmitoylation of STING (mouse specific) | Haag et al., 2018 |
| H-151 |
|
Palmitoylation of STING | Haag et al., 2018 |
| BPK-25 |
|
Palmitoylation of STING | Vinogradova et al., 2020 |
| Palbociclib |
|
Oligomerization of STING | Gao et al., 2022 |
| Agonists of STING | |||
| DMXAA |
|
Stability of STING closed conformation (mouse specific) | Gao et al., 2013c |
| CMA |
|
STING-CDN binding pocket (mouse specific) | Cavlar et al., 2013 |
| diABZI |
|
Stability of STING closed conformation (human specific) | Ramanjulu et al., 2018 |
| Compound 24b |
|
Stability of STING closed conformation | Xi et al., 2020 |
| SR-717 |
|
Stability of STING closed conformation | Chin et al., 2020 |
| Triazole 40 |
|
Stability of STING closed conformation | Song et al., 2021 |
| MSA-2 |
|
Stability of STING closed conformation | Pan et al., 2020 |
| Compound 53 |
|
Oligomerization of STING (human specific) | Lu et al., 2022; Pryde et al., 2021 |
Among the drugs that have been developed, several have yielded encouraging results in various disease models. In terms of cGAS inhibitors, X6 is an optimized product of antimalarial drugs (AMDs) and shows excellent therapeutic effects in mouse models of myocarditis (An et al., 2018). Furthermore, Ru.521 shows superior anti-inflammatory effects in macrophages in mouse models of AGS (Vincent et al., 2017), and rescues heart function in cGAS overactivation-induced sepsis mice (Xu et al., 2020b). Aspirin can also inhibit cGAS-mediated autoimmunity in mouse models of AGS (Dai et al., 2019). In terms of STING inhibitors, nitro fatty acids (NO2-FAs) have been shown to inhibit the production of IFN-I in fibroblasts of SAVI patients (Hansen et al., 2018a). Palbociclib can reduce the inflammatory response and tissue damage in a DSS-induced mouse models of colitis (Vinogradova et al., 2020; Zhao et al., 2022). Thus, inhibitors targeting cGAS and STING deserve further study in the treatment of autoimmune inflammatory diseases caused by abnormal activation of the pathway.
Regarding STING agonists, a range of drugs have been shown to be effective in the treatment of cancer. Intravenous injection of diABZI enhances STING-mediated antitumor activity in mouse models of colon tumor (Ramanjulu et al., 2018). Compound 24b, an analogue of diABZI, is also therapeutically effective in mice with colon tumors (Xi et al., 2020). In polyomavirus middle T antigen (PyMT) tumors, DMAXX promotes the production of STING-mediated antitumor factors, disrupts the tumor vascular system, and mediates tumor cell death. In addition, cGAMP analogue SR-717 activates STING by maintaining its “closed conformation”, thereby effectively increasing the efficiency of antigen delivery by promoting CD8+ T cell and DC activation. SR-717 also promotes PD-L1 expression in a STING-dependent manner, showing great anticancer potential (Chin et al., 2020). Although many drugs targeting cGAS and STING have been developed or modified for better function in disease treatment, their possible side effects and combined application with other drugs (e.g., anticancer drugs) deserve further study.
CONCLUSIONS AND FUTURE PERSPECTIVES
Since the discovery of STING in 2008 (Ishikawa & Barber, 2008; Zeng & Chen, 2008; Zhong et al., 2008) and cGAS in 2012 (Sun et al., 2013), our understanding of the cGAS-STING signaling pathway has greatly evolved. Unlike RNA receptors, which can distinguish non-self RNA from self-RNA, e.g., retinoic acid-inducible gene I (RIG-I) only recognizes 5’-(P)PP containing double-strand RNA with no ribose 2’-O-methylation (Rehwinkel & Gack, 2020), a feature shared by viral RNAs, cGAS binds to dsDNA in a sequence- and structure-independent manner and can thus recognize both foreign and host DNA. Various studies have focused on the mechanisms involved in preventing recognition of cytosolic self-DNA by cGAS under normal conditions. One such mechanism is that the DNase localized in the cytosol digests leaked DNA to prevent its accumulation and activation of cGAS in healthy cells (Ahn and Barber, 2014). Another mechanism is compartmentalization, like restraining DNA in the mitochondria and nucleus of healthy cells and cGAS on the plasma membrane and nucleus. Plasma membrane-localized cGAS is important to avoid self-DNA detection (Barnett et al., 2019) and cGAS in the nucleus is strictly tethered to chromatin (Cao et al., 2020; Michalski et al., 2020). Recent studies have also shown that the hyperphosphorylation of cGAS can prevent DNA binding and liquid phase separation necessary for cGAS activation, thereby inhibiting cGAS during mitosis when the nuclear envelope disassembles and cytoplasmic cGAS meets nuclear DNA (Boyer et al., 2020; Kujirai et al., 2020). However, it remains unclear whether other regulatory mechanisms and post-translational modifications inhibit cGAS recognition of nuclear DNA during cell cycle transition. Furthermore, as most post-translational modifications are reversible, how different enzymes cooperate during transition is also unknown. Recent research showed that STING forms a liquid phase separation state in response to high intracellular cGAMP and sequesters STING and TBK1 in the ER, preventing activation of signaling downstream of Golgi trafficking, with Mn2+ and Zn2+ reported to promote this process (Yu et al., 2021). However, the detailed mechanisms by which cGAMP levels determine liquid phase separation of STING and whether other regulatory factors are also involved require more study. Many transcriptional subtypes of STING are widely expressed in human tissues and induced by viral infections to interact with STING and TBK1 and inhibit IFN-I induction (Wang et al., 2018b). Whether these STING variants also undergo phase separation or whether their existence affects STING phase separation requires further study. As liquid phase separation is important in the regulation of many cellular functions, including regulation of cGAS-STING, whether liquid phase separation is also involved in blocking or promoting interactions with downstream adaptors deserves further investigation.
Recent studies have also focused on metabolism-mediated regulation of cGAS-STING signaling. Increased lipid peroxidation caused by glutathione peroxidase 4 (GPX4) inactivation leads to STING carbonylation and blockade of ER to Golgi translocation (Jia et al., 2020), while decreased cholesterol levels in the ER membrane facilitate STING/TBK1 interaction and signal transduction (York et al., 2015). On the other hand, abnormal activation of cGAS-STING signaling can promote lipid biosynthesis and accumulation in immune cells, indicating that interactions exist between lipid metabolism and cGAS-STING signaling (Bai et al., 2021). A negative regulatory loop also exists between STING and polyunsaturated fatty acid (PUFA) metabolism. STING can promote PUFA desaturation, which, in turn, inhibits STING function and resolves antiviral inflammatory response (Vila et al., 2022). As the center for energy metabolism, mitochondria oxidize pyruvate, fatty acids, and amino acids to facilitate ATP production through oxidative phosphorylation. Determining whether intermediate products produced during these processes link mitochondria more to cGAS-STING signaling should help clarify our understanding of the interplay between this energy center and antipathogen/damage signaling.
Many drugs and inhibitors targeting cGAS and STING have been developed for the treatment of inflammatory-related diseases and tumors (Zhao et al., 2022). However, the “double-edged sword” effects of cGAS-STING signaling during tumorigenesis and tumor therapy limit their effectiveness and reliability (Klarquist et al., 2014; Li & Chen, 2018; Wang et al., 2017a). Furthermore, the exact mechanisms and potential spatiotemporal effects of different tumors and stages remain ambiguous. Combination therapy using anti-tumor drugs, such as well-known PD-1 and PDL-1 antibodies, with cGAS and STING inhibitors for better clinical effects is worthy of in-depth investigation (Sen et al., 2019). In addition, little is known regarding the mechanisms of cytosolic cGAS-STING-mediated cellular senescence. Targeting this important physiological process to treat aging-related diseases is a very promising field in the future.
Ongoing efforts to translate the cGAS-STING signaling pathway from animal models to clinical application are still required for an accurate understanding of the etiological role of this pathway in disease pathogenesis. A key challenge for the future is how to precisely regulate rather than over- or under-regulate the cGAS-STING pathway in clinical practice, and its precise delineation will be a great step forward in the treatment of cGAS-STING-related diseases. Whether cGAS serves as more than a DNA receptor, e.g., regulating development like RIG-I, is an interesting question worthy of further study (Lefkopoulos et al., 2020; Wang et al., 2022c).
Funding Statement
This work was supported by the Natural Science Foundation of Zhejiang Province (LY23C190002), National Natural Science Foundation of China (32173004), and Natural Science Foundation of Ningbo City (202003N4011)
Contributor Information
Li Nie, Email: nieli@nbu.edu.cn.
Jiong Chen, Email: chenjiong@nbu.edu.cn.
References
- Aarreberg LD, Esser-Nobis K, Driscoll C, Shuvarikov A, Roby JA, Gale M Jr Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS-STING. Molecular Cell. 2019;74(4):801–815.e6. doi: 10.1016/j.molcel.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Barber GN Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. Journal of Virology. 2014;88(10):5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A ReGLUation of cGAS. Nature Immunology. 2016;17(4):347–349. doi: 10.1038/ni.3397. [DOI] [PubMed] [Google Scholar]
- Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, et al ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. Journal of Experimental Medicine. 2018;215(11):2868–2886. doi: 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Barber GN Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Current Opinion in Immunology. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Ahn J, Son S, Oliveira SC, Barber GN STING-dependent signaling underlies IL-10 controlled inflammatory colitis. Cell Reports. 2017;21(13):3873–3884. doi: 10.1016/j.celrep.2017.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Xia TL, Konno H, Konno K, Ruiz P, Barber GN Inflammation-driven carcinogenesis is mediated through STING. Nature Communications. 2014;5:5166. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Saitoh T, Kawai T. 2012. Nucleic acids recognition by innate immunity. Uirusu, 62(1): 39–46. (in Japanese)
- Al-Chalabi A, Hardiman O The epidemiology of ALS: a conspiracy of genes, environment and time. Nature Reviews Neurology. 2013;9(11):617–628. doi: 10.1038/nrneurol.2013.203. [DOI] [PubMed] [Google Scholar]
- An J, Minie M, Sasaki T, Woodward JJ, Elkon KB Antimalarial drugs as immune modulators: new mechanisms for old drugs. Annual Review of Medicine. 2017;68:317–330. doi: 10.1146/annurev-med-043015-123453. [DOI] [PubMed] [Google Scholar]
- An J, Woodward JJ, Lai WN, Minie M, Sun XZ, Tanaka L, et al Inhibition of cyclic GMP-AMP synthase using a novel antimalarial drug derivative in Trex1-deficient mice. Arthritis & Rheumatology. 2018;70(11):1807–1819. doi: 10.1002/art.40559. [DOI] [PubMed] [Google Scholar]
- An J, Woodward JJ, Sasaki T, Minie M, Elkon KB Cutting edge: antimalarial drugs inhibit IFN-β production through blockade of cyclic GMP-AMP synthase-DNA interaction. The Journal of Immunology. 2015;194(9):4089–4093. doi: 10.4049/jimmunol.1402793. [DOI] [PubMed] [Google Scholar]
- Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Jan Drexler D, et al cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature. 2017;549(7672):394–398. doi: 10.1038/nature23890. [DOI] [PubMed] [Google Scholar]
- Apel F, Andreeva L, Knackstedt LS, Streeck R, Frese CK, Goosmann C, et al The cytosolic DNA sensor cGAS recognizes neutrophil extracellular traps. Science Signaling. 2021;14(673):eaax7942. doi: 10.1126/scisignal.aax7942. [DOI] [PubMed] [Google Scholar]
- Bai JL, Liu F cGAS-STING signaling and function in metabolism and kidney diseases. Journal of Molecular Cell Biology. 2021;13(10):728–738. doi: 10.1093/jmcb/mjab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai JL, Liu F Nuclear cGAS: sequestration and beyond. Protein & Cell. 2022;13(2):90–101. doi: 10.1007/s13238-021-00869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, et al Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell. 2019;176(6):1432–1446.e11. doi: 10.1016/j.cell.2019.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum R, Sharma S, Organ JM, Jakobs C, Hornung V, Burr DB, et al STING contributes to abnormal bone formation induced by deficiency of DNase II in mice. Arthritis & Rheumatology. 2017;69(2):460–471. doi: 10.1002/art.39863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart DC, Sandborn WJ Crohn's disease. The Lancet. 2012;380(9853):1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG Protein arginine methylation in mammals: who, what, and why. Molecular Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JL, Sumpter R Jr, Levine B, Hooper LV Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host & Microbe. 2013;13(6):723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerzoug S, Rose S, Bounab B, Gosset D, Duneau L, Chenuet P, et al STING-dependent sensing of self-DNA drives silica-induced lung inflammation. Nature Communications. 2018;9(1):5226. doi: 10.1038/s41467-018-07425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot JM, Drouet L, Lioté F Kawasaki-like diseases and thrombotic coagulopathy in COVID-19: delayed over-activation of the STING pathway? Emerging Microbes & Infections. 2020a;9(1):1514–1522. doi: 10.1080/22221751.2020.1785336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot JM, Lioté F COVID-19 as a STING disorder with delayed over-secretion of interferon-beta. eBioMedicine. 2020;56:102801. doi: 10.1016/j.ebiom.2020.102801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc RS, Richard S Arginine methylation: the coming of age. Molecular Cell. 2017;65(1):8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Berthelot JM, Lioté F, Maugars Y, Sibilia J Lymphocyte changes in severe COVID-19: delayed over-activation of STING? Frontiers in Immunology. 2020b;11:607069. doi: 10.3389/fimmu.2020.607069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodda C, Reinert LS, Fruhwürth S, Richardo T, Sun CL, Zhang BC, et al HSV1 VP1–2 deubiquitinates STING to block type I interferon expression and promote brain infection. Journal of Experimental Medicine. 2020;217(7):e20191422. doi: 10.1084/jem.20191422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JA, Spangler CJ, Strauss JD, Cesmat AP, Liu PD, Mcginty RK, et al Structural basis of nucleosome-dependent cGAS inhibition. Science. 2020;370(6515):450–454. doi: 10.1126/science.abd0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Barlowe C Organization of the ER-Golgi interface for membrane traffic control. Nature Reviews Molecular Cell Biology. 2013;14(6):382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nature Reviews Immunology. 2016;16(7):407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478(7370):515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J Aging, cellular senescence, and cancer. Annual Review of Physiology. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canesso MCC, Lemos L, Neves TC, Marim FM, Castro TBR, Veloso ÉS, et al The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal Immunology. 2018;11(3):820–834. doi: 10.1038/mi.2017.88. [DOI] [PubMed] [Google Scholar]
- Cao DF, Han XN, Fan XY, Xu RM, Zhang XZ Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Research. 2020;30(12):1088–1097. doi: 10.1038/s41422-020-00422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu EJ, Zheng RD, Zhangchen ZL, Zhong RL, Huang F, et al Traditional Chinese medicine Lingguizhugan decoction ameliorate HFD-induced hepatic-lipid deposition in mice by inhibiting STING-mediated inflammation in macrophages. Chinese Medicine. 2022;17(1):7. doi: 10.1186/s13020-021-00559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TY, Shao S, Li B, Jin L, Lei J, Qiao HJ, et al Up-regulation of interferon-inducible protein 16 contributes to psoriasis by modulating chemokine production in keratinocytes. Scientific Reports. 2016;6:25381. doi: 10.1038/srep25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda D, Gonzalez AJ, Alomari M, Tandon K, Zervos XB From hepatitis A to E: a critical review of viral hepatitis. World Journal of Gastroenterology. 2021;27(16):1691–1715. doi: 10.3748/wjg.v27.i16.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V Species-specific detection of the antiviral small-molecule compound CMA by STING. The EMBO Journal. 2013;32(10):1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen H, Zhang JM, Wang YM, Simoneau A, Yang H, et al cGAS suppresses genomic instability as a decelerator of replication forks. Science Advances. 2020;6(42):eabb8941. doi: 10.1126/sciadv.abb8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Pei RJ, Zhu WD, Zeng R, Wang Y, Wang YY, et al An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. The Journal of Immunology. 2014;192(3):1162–1170. doi: 10.4049/jimmunol.1300798. [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC Shaping the nuclear action of NF-κB. Nature Reviews Molecular Cell Biology. 2004;5(5):392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Chen MX, Meng QC, Qin YF, Liang PP, Tan P, He L, et al TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Molecular Cell. 2016a;64(1):105–119. doi: 10.1016/j.molcel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016b;533(7604):493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun LJ, Chen ZJ Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nature Immunology. 2016c;17(10):1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Chen SY, Liu YQ, Zhou HC Advances in the Development ubiquitin-specific peptidase (USP) inhibitors. International Journal of Molecular Sciences. 2021;22(9):4546. doi: 10.3390/ijms22094546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Chen YF Ubiquitination of cGAS by TRAF6 regulates anti-DNA viral innate immune responses. Biochemical and Biophysical Research Communications. 2019;514(3):659–664. doi: 10.1016/j.bbrc.2019.05.022. [DOI] [PubMed] [Google Scholar]
- Chen YF, Wang LF, Jin JL, Luan Y, Chen C, Li Y, et al p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. Journal of Experimental Medicine. 2017;214(4):991–1010. doi: 10.1084/jem.20161387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin EN, Yu CG, Vartabedian VF, Jia Y, Kumar M, Gamo AM, et al Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science. 2020;369(6506):993–999. doi: 10.1126/science.abb4255. [DOI] [PubMed] [Google Scholar]
- Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, et al HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. The EMBO Journal. 2016;35(13):1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L, Li CH, Li YX, Yu QY, Yu HS, Li CH, et al Perillaldehyde inhibition of cGAS reduces dsDNA-induced interferon response. Frontiers in Immunology. 2021;12:655637. doi: 10.3389/fimmu.2021.655637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, et al Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498(7454):332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AC, Cai HC, Li T, Franco LH, Li XD, Nair VR, et al. 2015. Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host & Microbe, 17(6): 820–828.
- Cong XY, Yuan ZL, Du YJ, Wu B, Lu DF, Wu XJ, et al Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins. Journal of Biological Chemistry. 2019;294(30):11420–11432. doi: 10.1074/jbc.RA119.007367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Gajewski TF Endogenous and pharmacologic targeting of the STING pathway in cancer immunotherapy. Cytokine. 2016;77:245–247. doi: 10.1016/j.cyto.2015.08.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DJ, Field RH, Williams DG, Baran M, Bowie AG, Cunningham C, et al DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia. 2015;63(5):812–825. doi: 10.1002/glia.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree BAC, Arnold DL, Chataway J, Chitnis T, Fox RJ, Pozo Ramajo A, et al Secondary progressive multiple sclerosis: new insights. Neurology. 2021;97(8):378–388. doi: 10.1212/WNL.0000000000012323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ Type I interferonopathies: mendelian type I interferon up-regulation. Current Opinion in Immunology. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Cui SF, Yu QY, Chu L, Cui Y, Ding M, Wang QY, et al Nuclear cGAS functions non-canonically to enhance antiviral immunity via recruiting methyltransferase prmt5. Cell Reports. 2020;33(10):108490. doi: 10.1016/j.celrep.2020.108490. [DOI] [PubMed] [Google Scholar]
- Cui Y, Yu H, Zheng X, Peng R, Wang Q, Zhou Y, et al SENP7 potentiates cGAS activation by relieving SUMO-mediated inhibition of cytosolic DNA sensing. PLoS Pathogens. 2017;13(1):e1006156. doi: 10.1371/journal.ppat.1006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Huang YJ, He XH, Zhao M, Wang XZ, Liu ZS, et al Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176(6):1447–1460.e14. doi: 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansako H, Imai H, Ueda Y, Satoh S, Shimotohno K, Kato N High-level expression of STING restricts susceptibility to HBV by mediating type III IFN induction. FASEB Bioadvances. 2019;1(2):67–80. doi: 10.1096/fba.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansako H, Ueda Y, Okumura N, Satoh S, Sugiyama M, Mizokami M, et al The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS Journal. 2016;283(1):144–156. doi: 10.1111/febs.13563. [DOI] [PubMed] [Google Scholar]
- D'Arcy MS Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biology International. 2019;43(6):582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SE, Khatua AK, Popik W Nucleosomal dsDNA stimulates APOL1 expression in human cultured podocytes by activating the cGAS/IFI16-STING signaling pathway. Scientific Reports. 2019;9(1):15485. doi: 10.1038/s41598-019-51998-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco F, Cutarelli A, Catoi AF, Uberti BD, Cuccaro B, Roperto S Bovine delta papillomavirus E5 oncoprotein negatively regulates the cGAS-STING signaling pathway in cattle in a spontaneous model of viral disease. Frontiers in Immunology. 2022;13:937736. doi: 10.3389/fimmu.2022.937736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaetano A, Solodka K, Zanini G, Selleri V, Mattioli AV, Nasi M, et al Molecular mechanisms of mtDNA-mediated inflammation. Cells. 2021;10(11):2898. doi: 10.3390/cells10112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decout A, Katz JD, Venkatraman S, Ablasser A The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nature Reviews Immunology. 2021;21(9):548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria O, De Gassart A, Coso S, Gestermann N, Di Domizio J, Flatz L, et al STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(50):15408–15413. doi: 10.1073/pnas.1512832112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Meng T, Chen L, Wei WY, Wang P The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduction and Targeted Therapy. 2020a;5(1):11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng LF, Liang H, Xu M, Yang XM, Burnette B, Arina A, et al STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41(5):843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XB, Yu XY, Pei JF. 2020b. Regulation of interferon production as a potential strategy for COVID-19 treatment. arXiv preprint arXiv: 2003.00751.
- Dewar JM, Walter JC Mechanisms of DNA replication termination. Nature Reviews Molecular Cell Biology. 2017;18(8):507–516. doi: 10.1038/nrm.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewi Pamungkas Putri D, Kawasaki T, Murase M, Sueyoshi T, Deguchi T, Ori D, et al PtdIns3P phosphatases MTMR3 and MTMR4 negatively regulate innate immune responses to DNA through modulating STING trafficking. Journal of Biological Chemistry. 2019;294(21):8412–8423. doi: 10.1074/jbc.RA118.005731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Cao XZ, Lu J, Huang B, Liu YJ, Kato N, et al Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. Journal of Hepatology. 2013;59(1):52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Dobbs N, Burnaevskiy N, Chen DD, Gonugunta VK, Alto NM, Yan N STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host & Microbe. 2015;18(2):157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggin E, Madhavan MV, Bikdeli B, Chuich T, Laracy J, Biondi-Zoccai G, et al Cardiovascular considerations for patients, health care workers, and health systems during the COVID-19 pandemic. Journal of the American College of Cardiology. 2020;75(18):2352–2371. doi: 10.1016/j.jacc.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du MJ, Chen ZJ DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361(6403):704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugger BN, Dickson DW Pathology of neurodegenerative diseases. Cold Spring Harbor Perspectives in Biology. 2017;9(7):a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler K, Vertegaal ACO SUMOylation-mediated regulation of cell cycle progression and cancer. Trends in Biochemical Sciences. 2015;40(12):779–793. doi: 10.1016/j.tibs.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev RI, Schulman BA, Peter M Protein neddylation: beyond cullin-RING ligases. Nature Reviews Molecular Cell Biology. 2015;16(1):30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun SL, Fernandez D, Weiss TM, Li LY STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell. 2019;178(2):290–301.e10. doi: 10.1016/j.cell.2019.05.036. [DOI] [PubMed] [Google Scholar]
- Ergun SL, Li LY Structural insights into STING signaling. Trends in Cell Biology. 2020;30(5):399–407. doi: 10.1016/j.tcb.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Fang CJ, Mo F, Liu L, Du J, Luo M, Men K, et al Oxidized mitochondrial DNA sensing by STING signaling promotes the antitumor effect of an irradiated immunogenic cancer cell vaccine. Cellular & Molecular Immunology. 2021;18(9):2211–2223. doi: 10.1038/s41423-020-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Tschachler E, Eckhart L. 2020. Cytosolic DNA sensing through cGAS and STING is inactivated by gene mutations in pangolins. Apoptosis, 25(7–8): 474–480.
- Fischer JC, Bscheider M, Eisenkolb G, Lin CC, Wintges A, Otten V, et al RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Science Translational Medicine. 2017;9(386):eaag2513. doi: 10.1126/scitranslmed.aag2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho A, Melchior F Sumoylation: a regulatory protein modification in health and disease. Annual Review of Biochemistry. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Foley JF Membrane fusion stimulates STING. Science Signaling. 2012;5(235):ec202. [Google Scholar]
- Galluzzi L, Green DR Autophagy-independent functions of the autophagy machinery. Cell. 2019;177(7):1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Bataller R Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141(5):1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao DX, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, et al Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(42):E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao DX, Wu JX, Wu YT, Du FH, Aroh C, Yan N, et al Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013a;341(6148):903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JN, Zheng MG, Wu XY, Zhang H, Su H, Dang YF, et al CDK inhibitor Palbociclib targets STING to alleviate autoinflammation. EMBO Reports. 2022;23(6):e53932. doi: 10.15252/embr.202153932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al Cyclic [G(2', 5')pA(3', 5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013b;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang WY, Dai PH, Serganov AA, et al Structure-function analysis of STING activation by c[G(2', 5')pA(3', 5')p] and targeting by antiviral DMXAA. Cell. 2013c;154(4):748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC, et al Multivalent microtubule recognition by tubulin tyrosine ligase-like family glutamylases. Cell. 2015;161(5):1112–1123. doi: 10.1016/j.cell.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Baskaran S, Schekman R, Hurley JH The protein-vesicle network of autophagy. Current Opinion in Cell Biology. 2014;29:18–24. doi: 10.1016/j.ceb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Modifiers of Huntington's Disease (GeM-HD) Consortium CAG repeat not polyglutamine length determines timing of huntington's disease onset. Cell. 2019;178(4):887–900.e14. doi: 10.1016/j.cell.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M, Lahaye X, Nadalin F, Nader GPF, Puig Lombardi E, Herve S, et al The N-Terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Reports. 2019;26(9):2377–2393.e13. doi: 10.1016/j.celrep.2019.01.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Lu LL, Zhou Y, Liu JY, Ma HY, Fu LH, et al The novel STING antagonist H151 ameliorates cisplatin-induced acute kidney injury and mitochondrial dysfunction. American Journal of Physiology-Renal Physiology. 2021;320(4):F608–F616. doi: 10.1152/ajprenal.00554.2020. [DOI] [PubMed] [Google Scholar]
- Gong Y, Li GW, Tao J, Wu NN, Kandadi MR, Bi YG, et al. 2020. Double knockout of Akt2 and AMPK accentuates high fat diet-induced cardiac anomalies through a cGAS-STING-mediated mechanism. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 1866(10): 165855.
- Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu JJ, Dobbs N, et al Trafficking-mediated STING degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Reports. 2017;21(11):3234–3242. doi: 10.1016/j.celrep.2017.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, Stetson DB Cutting edge: cGAS is required for lethal autoimmune disease in the trex1-deficient mouse model of aicardi-goutières syndrome. The Journal of Immunology. 2015;195(5):1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser I, Morel-Maroger L, Rivière Y, Guillon JC, Tovey MG, Woodrow D, et al Interferon-induced disease in mice and rats. Annals of the New York Academy of Sciences. 1980;350(1):12–20. doi: 10.1111/j.1749-6632.1980.tb20602.x. [DOI] [PubMed] [Google Scholar]
- Gu TL, Yu DD, Xu L, Yao YL, Yao YG Tupaia GBP1 Interacts with STING to initiate autophagy and restrict Herpes Simplex Virus type 1 infection. The Journal of Immunology. 2021;207(11):2673–2680. doi: 10.4049/jimmunol.2100325. [DOI] [PubMed] [Google Scholar]
- Gui X, Yang H, Li T, Tan XJ, Shi PQ, Li MH, et al Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Han YX, Zhao XS, Wang JH, Liu F, Xu CX, et al STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrobial Agents and Chemotherapy. 2015;59(2):1273–1281. doi: 10.1128/AAC.04321-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Tang LD, Shu SN, Sehgal M, Sheraz M, Liu BW, et al Activation of stimulator of interferon genes in hepatocytes suppresses the replication of hepatitis B virus. Antimicrobial Agents and Chemotherapy. 2017;61(10):e00771–17. doi: 10.1128/AAC.00771-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YY, Jiang F, Kong LL, Li BQ, Yang YL, Zhang L, et al Cutting edge: USP27X deubiquitinates and stabilizes the DNA sensor cGAS to regulate cytosolic DNA-mediated signaling. The Journal of Immunology. 2019;203(8):2049–2054. doi: 10.4049/jimmunol.1900514. [DOI] [PubMed] [Google Scholar]
- Guo YY, Jiang F, Kong LL, Wu HF, Zhang HH, Chen XR, et al OTUD5 promotes innate antiviral and antitumor immunity through deubiquitinating and stabilizing STING. Cellular and Molecular Immunology. 2021;18(8):1945–1955. doi: 10.1038/s41423-020-00531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, et al Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559(7713):269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- Hall J, Brault A, Vincent F, Weng S, Wang H, Dumlao D, et al Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS One. 2017;12(9):e0184843. doi: 10.1371/journal.pone.0184843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LL, Zheng Y, Deng J, Nan ML, Xiao Y, Zhuang MW, et al SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy. Journal of Medical Virology. 2022;94(11):5174–5188. doi: 10.1002/jmv.27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LL, Zhuang MW, Deng J, Zheng Y, Zhang J, Nan ML, et al SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. Journal of Medical Virology. 2021;93(9):5376–5389. doi: 10.1002/jmv.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YL, Chen L, Liu HW, Jin ZC, Wu YF, Wu YP, et al Airway epithelial cGAS is critical for induction of experimental allergic airway inflammation. The Journal of Immunology. 2020;204(6):1437–1447. doi: 10.4049/jimmunol.1900869. [DOI] [PubMed] [Google Scholar]
- Han ZJ, Feng YH, Gu BH, Li YM, Chen H The post-translational modification, SUMOylation, and cancer (Review) International Journal of Oncology. 2018;52(4):1081–1094. doi: 10.3892/ijo.2018.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AL, Buchan GJ, Rühl M, Mukai K, Salvatore SR, Ogawa E, et al Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proceedings of the National Academy of Sciences of the United States of America. 2018a;115(33):E7768–E7775. doi: 10.1073/pnas.1806239115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Hanson JE, Sheng M Microglia in Alzheimer's disease. The Journal of Cell Biology. 2018b;217(2):459–472. doi: 10.1083/jcb.201709069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Prabakaran T, Laustsen A, Jørgensen SE, Rahbæk SH, Jensen SB, et al Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. The EMBO Journal. 2014;33(15):1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova A, Erttmann SF, Raffi FAM, Schmalz AM, Resch U, Anugula S, et al DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42(2):332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- He J, Hao RD, Liu D, Liu X, Wu SS, Guo ST, et al Inhibition of Hepatitis B Virus replication by activation of the cGAS-STING pathway. Journal of General Virology. 2016;97(12):3368–3378. doi: 10.1099/jgv.0.000647. [DOI] [PubMed] [Google Scholar]
- Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, et al Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nature Immunology. 2012;13(8):737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Schubert M, Tijhuis AE, Requesens M, Roorda M, van den Brink A, et al cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature. 2022;607(7918):366–373. doi: 10.1038/s41586-022-04847-2. [DOI] [PubMed] [Google Scholar]
- Hong Z, Mei JH, Li CH, Bai GH, Maimaiti M, Hu HY, et al STING inhibitors target the cyclic dinucleotide binding pocket. Proceedings of the National Academy of Sciences of the United States of America. 2021;118(24):e2105465118. doi: 10.1073/pnas.2105465118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YJ, Wei Y, Lautrup S, Yang BM, Wang YM, Cordonnier S, et al NAD+ supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer's disease via cGAS-STING. Proceedings of the National Academy of Sciences of the United States of America. 2021;118(37):e2011226118. doi: 10.1073/pnas.2011226118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MJ, Zhou M, Bao XH, Pan D, Jiao M, Liu XJ, et al ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. The Journal of Clinical Investigation. 2021;131(3):e139333. doi: 10.1172/JCI139333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, et al Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45(3):555–569. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Huang L, Li LL, Lemos H, Chandler PR, Pacholczyk G, Baban B, et al Cutting edge: DNA sensing via the STING adaptor in myeloid dendritic cells induces potent tolerogenic responses. The Journal of Immunology. 2013;191(7):3509–3513. doi: 10.4049/jimmunol.1301419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Yao YC, Hou XL, Wei L, Rao YH, Su Y, et al Macrophage SCAP contributes to metaflammation and lean NAFLD by activating STING-NF-κB signaling pathway. Cellular and Molecular Gastroenterology and Hepatology. 2022;14(1):1–26. doi: 10.1016/j.jcmgh.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature Structural & Molecular Biology. 2012;19(7):728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Hwang ES Replicative senescence and senescence-like state induced in cancer-derived cells. Mechanisms of Ageing and Development. 2002;123(12):1681–1694. doi: 10.1016/S0047-6374(02)00102-1. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Rogowski K, van Dijk J Polyglutamylation: a fine-regulator of protein function? EMBO Reports. 2008;9(7):636–641. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauhari A, Baranov SV, Suofu Y, Kim J, Singh T, Yablonska S, et al Melatonin inhibits cytosolic mitochondrial DNA-induced neuroinflammatory signaling in accelerated aging and neurodegeneration. The Journal of Clinical Investigation. 2020;130(6):3124–3136. doi: 10.1172/JCI135026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia MT, Qin DH, Zhao CY, Chai L, Yu ZX, Wang WW, et al Redox homeostasis maintained by GPX4 facilitates STING activation. Nature Immunology. 2020;21(7):727–735. doi: 10.1038/s41590-020-0699-0. [DOI] [PubMed] [Google Scholar]
- Jiang S, Luo J, Zhang YW, Cao Q, Wang YN, Xia NW, et al The porcine and chicken innate DNA sensing cGAS-STING-IRF signaling axes exhibit differential species specificity. The Journal of Immunology. 2022;209(2):412–426. doi: 10.4049/jimmunol.2101212. [DOI] [PubMed] [Google Scholar]
- Jiang XF, Liu GP, Hu ZY, Chen GQ, Chen JQ, Lv ZB cGAMP inhibits tumor growth in colorectal cancer metastasis through the STING/STAT3 axis in a zebrafish xenograft model. Fish & Shellfish Immunology. 2019;95:220–226. doi: 10.1016/j.fsi.2019.09.075. [DOI] [PubMed] [Google Scholar]
- Johnson BM, Uchimura T, Gallovic MD, Thamilarasan M, Chou WC, Gibson SA, et al STING agonist mitigates experimental autoimmune encephalomyelitis by stimulating type I IFN-dependent and -independent immune-regulatory pathways. The Journal of Immunology. 2021;206(9):2015–2028. doi: 10.4049/jimmunol.2001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis M, Philpott DJ, Ferrero RL Mammalian NLR proteins; discriminating foe from friend. Immunology and Cell Biology. 2007;85(6):495–502. doi: 10.1038/sj.icb.7100105. [DOI] [PubMed] [Google Scholar]
- Karimi-Googheri M, Daneshvar H, Khaleghinia M, Bidaki R, Arababadi MK Decreased expressions of STING but not IRF3 molecules in chronic HBV infected patients. Archives of Iranian Medicine. 2015;18(6):351–354. [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Lalonde K, Truesdell A, Gomes Welter P, Brocardo PS, Rosenstock TR, et al New avenues for the treatment of Huntington's disease. International Journal of Molecular Sciences. 2021;22(16):8363. doi: 10.3390/ijms22168363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Gupta R, Blanco LP, Yang ST, Shteinfer-Kuzmine A, Wang KN, et al VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019;366(6472):1531–1536. doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Klarquist J, Hennies CM, Lehn MA, Reboulet RA, Feau S, Janssen EM STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. The Journal of Immunology. 2014;193(12):6124–6134. doi: 10.4049/jimmunol.1401869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155(3):688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee ASY, Berger JM, Doudna JA Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Reports. 2013;3(5):1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujirai T, Zierhut C, Takizawa Y, Kim R, Negishi L, Uruma N, et al Structural basis for the inhibition of cGAS by nucleosomes. Science. 2020;370(6515):455–458. doi: 10.1126/science.abd0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X, Gentili M, Silvin A, Conrad C, Picard L, Jouve M, et al NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell. 2018;175(2):488–501.e22. doi: 10.1016/j.cell.2018.08.062. [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE Zinc finger proteins: new insights into structural and functional diversity. Current Opinion in Structural Biology. 2001;11(1):39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Lam E, Stein S, Falck-Pedersen E Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. Journal of Virology. 2014;88(2):974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama L, Adura C, Xie W, Tomita D, Kamei T, Kuryavyi V, et al Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nature Communications. 2019;10(1):2261. doi: 10.1038/s41467-019-08620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larabi A, Barnich N, Nguyen HTT New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16(1):38–51. doi: 10.1080/15548627.2019.1635384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrick JW, Mendelsohn AR Modulation of cGAS-STING pathway by nicotinamide riboside in Alzheimer's disease. Rejuvenation Research. 2021;24(5):397–402. doi: 10.1089/rej.2021.0062. [DOI] [PubMed] [Google Scholar]
- Lauterbach-Rivière L, Bergez M, Mönch S, Qu BQ, Riess M, Vondran FWR, et al Hepatitis B Virus DNA is a substrate for the cGAS/STING pathway but is not sensed in infected hepatocytes. Viruses. 2020;12(6):592. doi: 10.3390/v12060592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Woodruff TM TDP-43 puts the STING in ALS. Trends in Neurosciences. 2021;44(2):81–82. doi: 10.1016/j.tins.2020.12.001. [DOI] [PubMed] [Google Scholar]
- Lefkopoulos S, Polyzou A, Derecka M, Bergo V, Clapes T, Cauchy P, et al Repetitive elements trigger RIG-I-like receptor signaling that regulates the emergence of hematopoietic stem and progenitor cells. Immunity. 2020;53(5):934–951.e9. doi: 10.1016/j.immuni.2020.10.007. [DOI] [PubMed] [Google Scholar]
- Lemos H, Mohamed E, Huang L, Ou R, Pacholczyk G, Arbab AS, et al STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Research. 2016;76(8):2076–2081. doi: 10.1158/0008-5472.CAN-15-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepelley A, Della Mina E, Van Nieuwenhove E, Waumans L, Fraitag S, Rice GI, et al Enhanced cGAS-STING-dependent interferon signaling associated with mutations in ATAD3A. Journal of Experimental Medicine. 2021;218(10):e20201560. doi: 10.1084/jem.20201560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey AS, Titan SM, Powe NR, Coresh J, Inker LA Kidney disease, race, and GFR estimation. Clinical Journal of the American Society of Nephrology. 2020;15(8):1203–1212. doi: 10.2215/CJN.12791019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Zhang LL, Qian D, Cheng MX, Hu HY, Hong Z, et al RNF111-facilitated neddylation potentiates cGAS-mediated antiviral innate immune response. PLoS Pathogens. 2021a;17(3):e1009401. doi: 10.1371/journal.ppat.1009401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Hu L, Wang LW, Wang YX, Shao MQ, Chen YP, et al Iron activates cGAS-STING signaling and promotes hepatic inflammation. Journal of Agricultural and Food Chemistry. 2022a;70(7):2211–2220. doi: 10.1021/acs.jafc.1c06681. [DOI] [PubMed] [Google Scholar]
- Li QJ, Lin LB, Tong YL, Liu YT, Mou J, Wang XD, et al TRIM29 negatively controls antiviral immune response through targeting STING for degradation. Cell Discovery. 2018a;4:13. doi: 10.1038/s41421-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SL, Hong Z, Wang Z, Li F, Mei JH, Huang LL, et al The cyclopeptide Astin C specifically inhibits the innate immune CDN sensor STING. Cell Reports. 2018b;25(12):3405–3421.e7. doi: 10.1016/j.celrep.2018.11.097. [DOI] [PubMed] [Google Scholar]
- Li SR, Mirlekar B, Johnson BM, Brickey WJ, Wrobel JA, Yang N, et al STING-induced regulatory B cells compromise NK function in cancer immunity. Nature. 2022b;610(7931):373–380. doi: 10.1038/s41586-022-05254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen ZJ The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. Journal of Experimental Medicine. 2018;215(5):1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang TZ, Du MJ, Chen X, Du FH, Ren JY, et al Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science. 2021b;371(6535):eabc5386. doi: 10.1126/science.abc5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shu C, Yi GH, Chaton CT, Shelton CL, Diao JS, et al Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39(6):1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Zhu YY, Zhang X, An X, Weng MJ, Shi JQ, et al An alternatively spliced STING isoform localizes in the cytoplasmic membrane and directly senses extracellular cGAMP. The Journal of Clinical Investigation. 2022c;132(3):e144339. doi: 10.1172/JCI144339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Li XJ, Xie C, Cai SH, Li MQ, Jin HP, et al cGAS guards against chromosome end-to-end fusions during mitosis and facilitates replicative senescence. Protein & Cell. 2022d;13(1):47–64. doi: 10.1007/s13238-021-00879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL, Yu Z, Fang Q, Yang MJ, Huang JY, Li Z, et al The transmembrane endoplasmic reticulum-associated E3 ubiquitin ligase TRIM13 restrains the pathogenic-DNA-triggered inflammatory response. Science Advances. 2022e;8(4):496:eabh0496. doi: 10.1126/sciadv.abh0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YS, Shi F, Hu JM, Xie LL, Bode AM, Cao Y The role of deubiquitinases in oncovirus and host interactions. Journal of Oncology. 2019;2019:2128410. doi: 10.1155/2019/2128410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZX, Liu G, Sun LW, Teng Y, Guo XJ, Jia JH, et al PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathogens. 2015;11(3):e1004783. doi: 10.1371/journal.ppat.1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JQ, Hong Z, Sun BY, Guo ZX, Wang C, Zhu JJ The alternatively spliced isoforms of key molecules in the cGAS-STING signaling pathway. Frontiers in Immunology. 2021;12:771744. doi: 10.3389/fimmu.2021.771744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, et al Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host & Microbe. 2014;15(2):228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ Palmitoylation: policing protein stability and traffic. Nature Reviews Molecular Cell Biology. 2007;8(1):74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Linnerbauer M, Wheeler MA, Quintana FJ Astrocyte crosstalk in CNS inflammation. Neuron. 2020;108(4):608–622. doi: 10.1016/j.neuron.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio CWJ, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, et al cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. Journal of Virology. 2016;90(17):7789–7797. doi: 10.1128/JVI.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HP, Zhang HP, Wu XY, Ma DP, Wu JH, Wang L, et al Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018a;563(7729):131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- Liu SD, Chen SS, Li XR, Wu SY, Zhang Q, Jin QH, et al Lck/Hck/Fgr-mediated tyrosine phosphorylation negatively regulates TBK1 to restrain innate antiviral responses. Cell Host & Microbe. 2017a;21(6):754–768.e5. doi: 10.1016/j.chom.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Liu SH, Zhao KT, Su X, Lu L, Zhao H, Zhang XW, et al MITA/STING and its alternative splicing isoform MRP restrict Hepatitis B Virus replication. PLoS One. 2017b;12(1):e0169701. doi: 10.1371/journal.pone.0169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SQ, Cai X, Wu JX, Cong Q, Chen X, Li T, et al Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015a;347(6227):aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu XJ, Pu Y, Cron K, Deng LF, Kline J, Frazier WA, et al CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nature Medicine. 2015b;21(10):1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gordesky-Gold B, Leney-Greene M, Weinbren NL, Tudor M, Cherry S Inflammation-induced, STING-dependent autophagy restricts Zika Virus infection in the Drosophila brain. Cell Host & Microbe. 2018b;24(1):57–68.e3. doi: 10.1016/j.chom.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Sanchez GAM, et al Activated STING in a vascular and pulmonary syndrome. New England Journal of Medicine. 2014;371(6):507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu PB, Rivara S, Liu C, Ricci J, Ren XF, et al Clathrin-associated AP-1 controls termination of STING signalling. Nature. 2022;610(7933):761–767. doi: 10.1038/s41586-022-05354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Li JH, Chen JL, Li YM, Wang WX, Du XT, et al Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. Journal of Virology. 2015c;89(4):2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZF, Ji JF, Jiang XF, Shao T, Fan DD, Jiang XH, et al Characterization of cGAS homologs in innate and adaptive mucosal immunities in zebrafish gives evolutionary insights into cGAS-STING pathway. The FASEB Journal. 2020;34(6):7786–7809. doi: 10.1096/fj.201902833R. [DOI] [PubMed] [Google Scholar]
- Liu ZS, Zhang ZY, Cai H, Zhao M, Mao J, Dai J, et al RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell & Bioscience. 2018c;8:35. doi: 10.1186/s13578-018-0233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TM, Miyata K, Tanaka Y, Takahashi A Cellular senescence and senescence-associated secretory phenotype via the cGAS-STING signaling pathway in cancer. Cancer Science. 2020;111(2):304–311. doi: 10.1111/cas.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DF, Shang GJ, Li J, Lu Y, Bai XC, Zhang XW Activation of STING by targeting a pocket in the transmembrane domain. Nature. 2022;604(7906):557–562. doi: 10.1038/s41586-022-04559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Wang H, Wang ZH, Cai HC, Lu ZG, Li Y, et al A STING-activating nanovaccine for cancer immunotherapy. Nature Nanotechnology. 2017;12(7):648–654. doi: 10.1038/nnano.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo WW, Li S, Li C, Lian H, Yang Q, Zhong B, et al iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nature Immunology. 2016;17(9):1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- Luo XJ, Li HG, Ma LQ, Zhou J, Guo X, Woo SL, et al Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology. 2018;155(6):1971–1984.e4. doi: 10.1053/j.gastro.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther J, Khan S, Gala MK, Kedrin D, Sridharan G, Goodman RP, et al Hepatic gap junctions amplify alcohol liver injury by propagating cGAS-mediated IRF3 activation. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(21):11667–11673. doi: 10.1073/pnas.1911870117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DP, Yang M, Wang QS, Sun CY, Shi HB, Jing WQ, et al Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Science Advances. 2021;7(13):eabc1834. doi: 10.1126/sciadv.abc1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jacobs SR, West JA, Stopford C, Zhang ZG, Davis Z, et al Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(31):E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Inoue T, Ouchi H, Jao TM, Inoue R, Nishi H, et al Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Reports. 2019;29(5):1261–1273.e6. doi: 10.1016/j.celrep.2019.09.050. [DOI] [PubMed] [Google Scholar]
- Maiuri T, Hung CLK, Suart C, Begeja N, Barba-Bazan C, Peng Y, et al DNA repair in huntington's disease and spinocerebellar ataxias: somatic instability and alternative hypotheses. Journal of Huntington’s Disease. 2021;10(1):165–173. doi: 10.3233/JHD-200414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, Ma A Ubiquitin makes its mark on immune regulation. Immunity. 2010;33(6):843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Luo W, Zhang L, Wu WW, Yuan LS, Xu H, et al STING-IRF3 triggers endothelial inflammation in response to free fatty acid-induced mitochondrial damage in diet-induced obesity. Arteriosclerosis, Thrombosis, and Vascular Biology. 2017;37(5):920–929. doi: 10.1161/ATVBAHA.117.309017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR, Blomquist CM, Henare KL, Jirik FR Stimulator of interferon genes (STING) activation exacerbates experimental colitis in mice. Scientific Reports. 2019;9(1):14281. doi: 10.1038/s41598-019-50656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Burai R, Vest RT, Bonanno LN, Lehallier B, Zardeneta ME, et al Activation of the STING-dependent type I interferon response reduces microglial reactivity and neuroinflammation. Neuron. 2017;96(6):1290–1302.e6. doi: 10.1016/j.neuron.2017.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, et al BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science. 2018;359(6378):eaao6047. doi: 10.1126/science.aao6047. [DOI] [PubMed] [Google Scholar]
- Mdkhana B, Askari NSS, Ramakrishnan RK, Goel S, Hamid Q, Halwani R Nucleic acid-sensing pathways during SARS-CoV-2 infection: expectations versus reality. Journal of Inflammation Research. 2021;14:199–216. doi: 10.2147/JIR.S277716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski S, de Oliveira Mann CC, Stafford CA, Witte G, Bartho J, Lammens K, et al Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature. 2020;587(7835):678–682. doi: 10.1038/s41586-020-2748-0. [DOI] [PubMed] [Google Scholar]
- Min JR, Liu K Structures of chromatin modulators in complex with nucleosome. Current Opinion in Chemical Biology. 2021;63:105–114. doi: 10.1016/j.cbpa.2021.02.018. [DOI] [PubMed] [Google Scholar]
- Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, et al STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171(4):809–823.e13. doi: 10.1016/j.cell.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata M, Kosaka A, Yajima Y, Yasuda S, Ohara M, Ohara K, et al A critical role of STING-triggered tumor-migrating neutrophils for anti-tumor effect of intratumoral cGAMP treatment. Cancer Immunology, Immunotherapy. 2021;70(8):2301–2312. doi: 10.1007/s00262-021-02864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M, Gombault A, Lacerda-Queiroz N, Panek C, Savigny F, Sbeity M, et al Self-DNA release and STING-dependent sensing drives inflammation to cigarette smoke in mice. Scientific Reports. 2019;9(1):14848. doi: 10.1038/s41598-019-51427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni GX, Konno H, Barber GN Ubiquitination of STING at lysine 224 controls IRF3 activation. Science Immunology. 2017;2(11):eaah7119. doi: 10.1126/sciimmunol.aah7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni GX, Ma Z, Wong JP, Zhang ZG, Cousins E, Major MB, et al PPP6C negatively regulates STING-dependent innate immune responses. mBio. 2020;11(4):e01728–20. doi: 10.1128/mBio.01728-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai CJ, Wolf N, Chang IC, Kirn G, Marcus A, Ndubaku CO, et al NK cells mediate clearance of CD8+ T cell-resistant tumors in response to STING agonists. Science Immunology. 2020;5(45):eaaz2738. doi: 10.1126/sciimmunol.aaz2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning XH, Wang YT, Jing M, Sha MY, Lv MZ, Gao PF, et al Apoptotic caspases suppress type I interferon production via the cleavage of cGAS, MAVS, and IRF3. Molecular Cell. 2019;74(1):19–31.e7. doi: 10.1016/j.molcel.2019.02.013. [DOI] [PubMed] [Google Scholar]
- Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, et al Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57(1):46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Mukai K, Saito K, Arai H, Taguchi T The binding of TBK1 to STING requires exocytic membrane traffic from the ER. Biochemical and Biophysical Research Communications. 2018;503(1):138–145. doi: 10.1016/j.bbrc.2018.05.199. [DOI] [PubMed] [Google Scholar]
- Okin D, Medzhitov R Evolution of inflammatory diseases. Current Biology. 2012;22(17):R733–R740. doi: 10.1016/j.cub.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M, Rodrigues DR, Guillory V, Kut E, Giotis ES, Skinner MA, et al Chicken cGAS senses fowlpox virus infection and regulates macrophage effector functions. Frontiers in Immunology. 2021;11:613079. doi: 10.3389/fimmu.2020.613079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ Ulcerative colitis. The Lancet. 2012;380(9853):1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- Ouyang SY, Song XQ, Wang YY, Ru H, Shaw N, Jiang Y, et al Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36(6):1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijo J, Döring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, et al CGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathogens. 2016;12(4):e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan BS, Perera SA, Piesvaux JA, Presland JP, Schroeder GK, Cumming JN, et al An orally available non-nucleotide STING agonist with antitumor activity. Science. 2020;369(6506):eaba6098. doi: 10.1126/science.aba6098. [DOI] [PubMed] [Google Scholar]
- Park Y, Jin HS, Aki D, Lee J, Liu YC The ubiquitin system in immune regulation. Advances in Immunology. 2014;124:17–66. doi: 10.1016/B978-0-12-800147-9.00002-9. [DOI] [PubMed] [Google Scholar]
- Parmar JJ, Padinhateeri R Nucleosome positioning and chromatin organization. Current Opinion in Structural Biology. 2020;64:111–118. doi: 10.1016/j.sbi.2020.06.021. [DOI] [PubMed] [Google Scholar]
- Pathare GR, Decout A, Glück S, Cavadini S, Makasheva K, Hovius R, et al Structural mechanism of cGAS inhibition by the nucleosome. Nature. 2020;587(7835):668–672. doi: 10.1038/s41586-020-2750-6. [DOI] [PubMed] [Google Scholar]
- Paul BD, Snyder SH, Bohr VA Signaling by cGAS-STING in neurodegeneration, neuroinflammation, and aging. Trends in Neurosciences. 2021;44(2):83–96. doi: 10.1016/j.tins.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, et al STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(41):16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokatayev V, Yang K, Tu XT, Dobbs N, Wu JJ, Kalb RG, et al Homeostatic regulation of STING protein at the resting state by stabilizer TOLLIP. Nature Immunology. 2020;21(2):158–167. doi: 10.1038/s41590-019-0569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun CL, et al Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. The EMBO Journal. 2018;37(8):e97858. doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde DC, Middya S, Banerjee M, Shrivastava R, Basu S, Ghosh R, et al The discovery of potent small molecule activators of human STING. European Journal of Medicinal Chemistry. 2021;209:112869. doi: 10.1016/j.ejmech.2020.112869. [DOI] [PubMed] [Google Scholar]
- Qiao JT, Cui C, Qing L, Wang LS, He TY, Yan F, et al Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism. 2018;81:13–24. doi: 10.1016/j.metabol.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathogens. 2014;10(9):e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabellino A, Andreani C, Scaglioni PP The role of PIAS SUMO E3-Ligases in cancer. Cancer Research. 2017;77(7):1542–1547. doi: 10.1158/0008-5472.CAN-16-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Peter M Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series. EMBO Reports. 2008;9(10):969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanjulu JM, Pesiridis GS, Yang JS, Concha N, Singhaus R, Zhang SY, et al Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564(7736):439–443. doi: 10.1038/s41586-018-0705-y. [DOI] [PubMed] [Google Scholar]
- Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, et al HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503(7476):402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski MJ Cellular senescence: what, why, and how. Wounds. 2017;29(6):168–174. [PubMed] [Google Scholar]
- Rehwinkel J, Gack MU RIG-I-like receptors: their regulation and roles in RNA sensing. Nature Reviews:Immunology. 2020;20(9):537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DS, Lucchinetti CF, Calabresi PA Multiple sclerosis. New England Journal of Medicine. 2018;378(2):169–180. doi: 10.1056/NEJMra1401483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-García E, Olagüe C, Ríus-Rocabert S, Ferrero R, Llorens C, Larrea E, et al TMEM173 alternative spliced isoforms modulate viral replication through the STING pathway. Immunohorizons. 2018;2(11):363–376. doi: 10.4049/immunohorizons.1800068. [DOI] [PubMed] [Google Scholar]
- Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, et al A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143(4):564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Rui YJ, Su JM, Shen S, Hu Y, Huang DB, Zheng WW, et al Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduction and Targeted Therapy. 2021;6(1):123. doi: 10.1038/s41392-021-00515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, et al The N-ethyl-N-nitrosourea-induced goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infection and Immunity. 2011;79(2):688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T, Rodriguez BL, Chen LM, Corte CMD, Morikawa N, Fujimoto J, et al Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discovery. 2019;9(5):646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nature Communications. 2018;9(1):613. doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Yang A, Tan B, Kim S, Liang QM, Choi Y, et al Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Reports. 2015;13(2):440–449. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang GJ, Zhang CG, Chen ZJ, Bai XC, Zhang XW Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature. 2019;567(7748):389–393. doi: 10.1038/s41586-019-0998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Rajendrarao S, Shahani N, Ramírez-Jarquín UN, Subramaniam S Cyclic GMP-AMP synthase promotes the inflammatory and autophagy responses in Huntington disease. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(27):15989–15999. doi: 10.1073/pnas.2002144117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CR, Yang XK, Liu Y, Li HP, Chu HY, Li GH, et al ZDHHC18 negatively regulates cGAS-mediated innate immunity through palmitoylation. The EMBO Journal. 2022;41(11):e109272. doi: 10.15252/embj.2021109272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HP, Wu JX, Chen ZJ, Chen C Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(29):8947–8952. doi: 10.1073/pnas.1507317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu T, Altman MD, Baltus GA, Childers M, Ellis JM, Gunaydin H, et al Discovery of a novel cGAMP competitive ligand of the inactive form of STING. ACS Medicinal Chemistry Letters. 2019;10(1):92–97. doi: 10.1021/acsmedchemlett.8b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, et al Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561(7722):258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder NA, Silva GM Deubiquitinating enzymes (DUBs): regulation, homeostasis, and oxidative stress response. Journal of Biological Chemistry. 2021;297(3):101077. doi: 10.1016/j.jbc.2021.101077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Luo ZQ Post-translational regulation of ubiquitin signaling. Journal of Cell Biology. 2019;218(6):1776–1786. doi: 10.1083/jcb.201902074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Ma FL, Herrup K Accumulation of cytoplasmic DNA due to ATM deficiency activates the microglial viral response system with neurotoxic consequences. Journal of Neuroscience. 2019;39(32):6378–6394. doi: 10.1523/JNEUROSCI.0774-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZL, Wang XY, Zhang Y, Gu WT, Shen AC, Ding CY, et al Structure-activity relationship study of amidobenzimidazole analogues leading to potent and systemically administrable stimulator of interferon gene (STING) agonists. Journal of Medicinal Chemistry. 2021;64(3):1649–1669. doi: 10.1021/acs.jmedchem.0c01900. [DOI] [PubMed] [Google Scholar]
- Song ZM, Lin H, Yi XM, Guo W, Hu MM, Shu HB KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(35):21568–21575. doi: 10.1073/pnas.1922330117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger HG, Macvicar T, Bahat A, Fiedler KU, Hermans S, Ehrentraut D, et al Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity. Nature Metabolism. 2021;3(5):636–650. doi: 10.1038/s42255-021-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Woo JS, Wu BB, El-Sherbiny YM, Leung J, Chupradit K, et al The Ca2+ sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nature Immunology. 2019;20(2):152–162. doi: 10.1038/s41590-018-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhagen F, Zillinger T, Peukert K, Fox M, Thudium M, Barchet W, et al Suppressive oligodeoxynucleotides containing TTAGGG motifs inhibit cGAS activation in human monocytes. European Journal of Immunology. 2018;48(4):605–611. doi: 10.1002/eji.201747338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JM, Shen S, Hu Y, Chen SQ, Cheng LY, Cai Y, et al. 2022. SARS-CoV-2 ORF3a inhibits cGAS-STING-mediated autophagy flux and antiviral function. Journal of Medical Virology,doi: 10.1002/jmv.28175.
- Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, et al USP13 negatively regulates antiviral responses by deubiquitinating STING. Nature Communications. 2017;8:15534. doi: 10.1038/ncomms15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LJ, Wu JX, Du FH, Chen X, Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WX, Li Y, Chen L, Chen HH, You FP, Zhou X, et al ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(21):8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XN, Liu T, Zhao J, Xia HS, Xie J, Guo Y, et al DNA-PK deficiency potentiates cGAS-mediated antiviral innate immunity. Nature Communications. 2020;11(1):6182. doi: 10.1038/s41467-020-19941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Lio CJ, Campeau A, Steger M, Ay F, Mann M, et al The tumor suppressor kinase DAPK3 drives tumor-intrinsic immunity through the STING-IFN-β pathway. Nature Immunology. 2021;22(4):485–496. doi: 10.1038/s41590-021-00896-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Wu B, Chen TT, Fan C, Zhao JN, Xiong CD, et al Synthesis and pharmacological evaluation of tetrahydro-γ-carboline derivatives as potent anti-inflammatory agents targeting cyclic GMP-AMP synthase. Journal of Medicinal Chemistry. 2021;64(11):7667–7690. doi: 10.1021/acs.jmedchem.1c00398. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science Signaling. 2012;5(214):ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao JL, Zhou X, Jiang ZF cGAS-cGAMP-STING: the three musketeers of cytosolic DNA sensing and signaling. IUBMB Life. 2016;68(11):858–870. doi: 10.1002/iub.1566. [DOI] [PubMed] [Google Scholar]
- Tian MF, Liu WY, Zhang Q, Huang YQ, Li W, Wang WB, et al MYSM1 represses innate immunity and autoimmunity through suppressing the cGAS-STING pathway. Cell Reports. 2020;33(3):108297. doi: 10.1016/j.celrep.2020.108297. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33(5):765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Tu DQ, Zhu ZH, Zhou AY, Yun CH, Lee KE, Toms AV, et al Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Reports. 2013;3(3):747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, et al PINK1 mutations are associated with sporadic early-onset parkinsonism. Annals of Neurology. 2004;56(3):336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- Vane JR, Botting RM. 2003. The mechanism of action of aspirin. Thrombosis Research, 110(5–6): 255–258.
- Vanpouille-Box C, Formenti SC, Demaria S TREX1 dictates the immune fate of irradiated cancer cells. OncoImmunology. 2017;6(9):e1339857. doi: 10.1080/2162402X.2017.1339857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A The ubiquitin system, autophagy, and regulated protein degradation. Annual Review of Biochemistry. 2017;86:123–128. doi: 10.1146/annurev-biochem-061516-044859. [DOI] [PubMed] [Google Scholar]
- Verrier ER, Yim SA, Heydmann L, El Saghire H, Bach C, Turon-Lagot V, et al Hepatitis B Virus evasion from cyclic guanosine monophosphate-adenosine monophosphate synthase sensing in human hepatocytes. Hepatology. 2018;68(5):1695–1709. doi: 10.1002/hep.30054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila IK, Chamma H, Steer A, Saccas M, Taffoni C, Turtoi E, et al STING orchestrates the crosstalk between polyunsaturated fatty acid metabolism and inflammatory responses. Cell Metabolism. 2022;34(1):125–139.e8. doi: 10.1016/j.cmet.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J, Adura C, Gao P, Luz A, Lama L, Asano Y, et al Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nature Communications. 2017;8(1):750. doi: 10.1038/s41467-017-00833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova EV, Zhang XY, Remillard D, Lazar DC, Suciu RM, Wang YJ, et al An activity-guided map of electrophile-cysteine interactions in primary human T cells. Cell. 2020;182(4):1009–1026.e29. doi: 10.1016/j.cell.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Cambier S, Gray EE, Stetson DB Tight nuclear tethering of cGAS is essential for preventing autoreactivity. eLife. 2019;8:e47491. doi: 10.7554/eLife.47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sun ZY, Zhao CX, Zhang ZW, Wang HR, Liu Y, et al Maintaining manganese in tumor to activate cGAS-STING pathway evokes a robust abscopal anti-tumor effect. Journal of Controlled Release. 2021a;331:480–490. doi: 10.1016/j.jconrel.2021.01.036. [DOI] [PubMed] [Google Scholar]
- Wang CY, Sharma N, Veleeparambil M, Kessler PM, Willard B, Sen GC STING-mediated interferon induction by Herpes Simplex Virus 1 requires the protein tyrosine kinase Syk. mBio. 2021b;12(6):e0322821. doi: 10.1128/mbio.03228-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Wang X, Veleeparambil M, Kessler PM, Willard B, Chattopadhyay S, et al EGFR-mediated tyrosine phosphorylation of STING determines its trafficking route and cellular innate immunity functions. The EMBO Journal. 2020a;39(22):e104106. doi: 10.15252/embj.2019104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu SQ, Chen X, Shi HP, Chen C, Sun LJ, et al cGAS is essential for the antitumor effect of immune checkpoint blockade. Proceedings of the National Academy of Sciences of the United States of America. 2017a;114(7):1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang S, Liu L, Wang H, Yang B HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Research. 2017b;232:13–21. doi: 10.1016/j.virusres.2017.01.016. [DOI] [PubMed] [Google Scholar]
- Wang MD, Sooreshjani MA, Mikek C, Opoku-Temeng C, Sintim HO Suramin potently inhibits cGAMP synthase, cGAS, in THP1 cells to modulate IFN-β levels. Future Medicinal Chemistry. 2018a;10(11):1301–1317. doi: 10.4155/fmc-2017-0322. [DOI] [PubMed] [Google Scholar]
- Wang PH, Fung SY, Gao WW, Deng JJ, Cheng Y, Chaudhary V, et al A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing. Nucleic Acids Research. 2018b;46(8):4054–4071. doi: 10.1093/nar/gky186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang LY, Hong Z, Lv ZS, Mao ZM, Tang YJ, et al The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathogens. 2017c;13(3):e1006264. doi: 10.1371/journal.ppat.1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu X, Cui Y, Tang YJ, Chen W, Li SL, et al The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41(6):919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang C, Wang XJ, Miao JM, Chen WT, Zhou YR, et al. 2022a. Driving axon regeneration by orchestrating neuronal and non-neuronal innate immune responses via the IFNγ-cGAS-STING axis. Neuron,doi: 10.1016/j.neuron.2022.10.028.
- Wang XX, Rao HY, Zhao JM, Wee A, Li XH, Fei R, et al STING expression in monocyte-derived macrophages is associated with the progression of liver inflammation and fibrosis in patients with nonalcoholic fatty liver disease. Laboratory Investigation. 2020b;100(4):542–552. doi: 10.1038/s41374-019-0342-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qian G, Zhu LY, Zhao Z, Liu YN, Han WD, et al HIV-1 Vif suppresses antiviral immunity by targeting STING. Cellular & Molecular Immunology. 2022b;19(1):108–121. doi: 10.1038/s41423-021-00802-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YG, Dasso M SUMOylation and deSUMOylation at a glance. Journal of Cell Science. 2009;122(23):4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Lian QS, Yang B, Yan SS, Zhou HY, He L, et al TRIM30α is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathogens. 2015;11(6):e1005012. doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT, Lu HJ, Fang CY, Xu J. 2020c. Palmitoylation as a signal for delivery. In: Xu J. Regulation of Cancer Immune Checkpoints. Singapore: Springer, 399–424.
- Wang YY, Nie L, Xu XX, Shao T, Fan DD, Lin AF, et al Essential role of RIG-I in hematopoietic precursor emergence in primitive hematopoiesis during zebrafish development. Immunohorizons. 2022c;6(5):283–298. doi: 10.4049/immunohorizons.2200028. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Chen Q, Zhu HM, Yin XN, Wang K, Liu YH, et al Enhancing the immune response and tumor suppression effect of antitumor vaccines adjuvanted with non-nucleotide small molecule STING agonist. Chinese Chemical Letters. 2021c;32(6):1888–1892. doi: 10.1016/j.cclet.2021.01.036. [DOI] [Google Scholar]
- Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, et al The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host & Microbe. 2015;17(6):811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, et al Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2017. Global Hepatitis Report, 2017. Geneva: World Health Organization.
- Wiens KE, Ernst JD The mechanism for Type I interferon induction by Mycobacterium tuberculosis is bacterial strain-dependent. PLoS Pathogens. 2016;12(8):e1005809. doi: 10.1371/journal.ppat.1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MYK, et al STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Li WW, Shao YM, Avey D, Fu BS, Gillen J, et al Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host & Microbe. 2015;18(3):333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Zhao L, Han BB, Hu HG, Zhang BD, Li WH, et al A novel STING agonist for cancer immunotherapy and a SARS-CoV-2 vaccine adjuvant. Chemical Communications. 2021a;57(4):504–507. doi: 10.1039/D0CC06959K. [DOI] [PubMed] [Google Scholar]
- Wu JX, Sun LJ, Chen X, Du FH, Shi HP, Chen C, et al Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XC, Wang ZY, Qiao D, Yuan Y, Han C, Yang N, et al Porcine circovirus type 2 infection attenuates the K63-linked ubiquitination of STING to inhibit IFN-β induction via p38-MAPK pathway. Veterinary Microbiology. 2021b;258:109098. doi: 10.1016/j.vetmic.2021.109098. [DOI] [PubMed] [Google Scholar]
- Wu XM, Wu FH, Wang XQ, Wang LL, Siedow JN, Zhang WG, et al Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Research. 2014;42(13):8243–8257. doi: 10.1093/nar/gku569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YK, Li ST Role of post-translational modifications of cGAS in innate immunity. International Journal of Molecular Sciences. 2020;21(21):7842. doi: 10.3390/ijms21217842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YK, Song K, Hao WZ, Li J, Wang LY, Li ST Nuclear soluble cGAS senses double-stranded DNA virus infection. Communications Biology. 2022;5(1):433. doi: 10.1038/s42003-022-03400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi QM, Wang MJ, Jia WQ, Yang MJ, Hu JP, Jin J, et al Design, synthesis, and biological evaluation of amidobenzimidazole derivatives as stimulator of interferon genes (STING) receptor agonists. Journal of Medicinal Chemistry. 2020;63(1):260–282. doi: 10.1021/acs.jmedchem.9b01567. [DOI] [PubMed] [Google Scholar]
- Xia PY, Ye BQ, Wang S, Zhu XX, Du Y, Xiong Z, et al Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nature Immunology. 2016;17(4):369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- Xia T, Yi XM, Wu X, Shang J, Shu HB PTPN1/2-mediated dephosphorylation of MITA/STING promotes its 20S proteasomal degradation and attenuates innate antiviral response. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(40):20063–20069. doi: 10.1073/pnas.1906431116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Lama L, Adura C, Tomita D, Glickman JF, Tuschl T, et al Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(24):11946–11955. doi: 10.1073/pnas.1905013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing JJ, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, et al TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nature Communications. 2017;8(1):945. doi: 10.1038/s41467-017-00101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DW, Tian YZ, Xia Q, Ke BB The cGAS-STING pathway: novel perspectives in liver diseases. Frontiers in Immunology. 2021;12:682736. doi: 10.3389/fimmu.2021.682736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yu DD, Peng L, Wu Y, Fan Y, Gu TL, et al An alternative splicing of Tupaia STING modulated anti-RNA virus responses by targeting MDA5-LGP2 and IRF3. Journal of Immunology. 2020a;204(12):3191–3204. doi: 10.4049/jimmunol.1901320. [DOI] [PubMed] [Google Scholar]
- Xu MM, Pu Y, Han DL, Shi YY, Cao XZ, Liang H, et al Dendritic cells but not macrophages sense tumor mitochondrial DNA for cross-priming through signal regulatory protein α signaling. Immunity. 2017;47(2):363–373.e5. doi: 10.1016/j.immuni.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu QQ, Xiong HL, Zhu WX, Liu YP, Du Y Small molecule inhibition of cyclic GMP-AMP synthase ameliorates sepsis-induced cardiac dysfunction in mice. Life Sciences. 2020b;260:118315. doi: 10.1016/j.lfs.2020.118315. [DOI] [PubMed] [Google Scholar]
- Yamashiro LH, Wilson SC, Morrison HM, Karalis V, Chung JYJ, Chen KJ, et al Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nature Communications. 2020;11(1):3382. doi: 10.1038/s41467-020-17156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Liu Y, Cui YH, Song D, Zhang G, Ma SJ, et al RNF90 negatively regulates cellular antiviral responses by targeting MITA for degradation. PLoS Pathogens. 2020;16(3):e1008387. doi: 10.1371/journal.ppat.1008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang HZ, Ren JY, Chen Q, Chen ZJ CGAS is essential for cellular senescence. Proceedings of the National Academy of Sciences of the United States of America. 2017a;114(23):E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Tang XS, Nandakumar KS, Cheng K Autophagy induced by STING, an unnoticed and primordial function of cGAS. Cellular & Molecular Immunology. 2019;16(8):683–684. doi: 10.1038/s41423-019-0240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KD, Huang QT, Wang RY, Zeng Y, Cheng MY, Xue Y, et al African swine fever virus MGF505–11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway. Veterinary Microbiology. 2021;263:109265. doi: 10.1016/j.vetmic.2021.109265. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang LL, Ketkar H, Ma JZ, Yang G, Cui S, et al UBXN3B positively regulates STING-mediated antiviral immune responses. Nature Communications. 2018;9(1):2329. doi: 10.1038/s41467-018-04759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YF, He Y, Wang XX, Liang ZW, He G, Zhang P, et al Protein SUMOylation modification and its associations with disease. Open Biology. 2017b;7(10):170167. doi: 10.1098/rsob.170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye LY, Zhang Q, Liuyu TZ, Xu ZG, Zhang MX, Luo MH, et al USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PLoS Pathogens. 2019;15(4):e1007680. doi: 10.1371/journal.ppat.1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi GH, Wen YH, Shu C, Han QX, Konan KV, Li PW, et al Hepatitis C virus NS4B can suppress STING accumulation to evade innate immune responses. Journal of Virology. 2016;90(1):254–265. doi: 10.1128/JVI.01720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang XM, Tu DQ, Eck MJ, et al Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular Cell. 2012;46(6):735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T Viral RNA detection by RIG-I-like receptors. Current Opinion in Immunology. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, et al Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163(7):1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CH, Davidson S, Harapas CR, Hilton JB, Mlodzianoski MJ, Laohamonthonkul P, et al TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020a;183(3):636–649.e18. doi: 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Tian HB, Deng HY PPM1G restricts innate immune signaling mediated by STING and MAVS and is hijacked by KSHV for immune evasion. Science Advances. 2020b;6(47):eabd0276. doi: 10.1126/sciadv.abd0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XY, Zhang LY, Shen JX, Zhai YF, Jiang QF, Yi MR, et al The STING phase-separator suppresses innate immune signalling. Nature Cell Biology. 2021;23(4):330–340. doi: 10.1038/s41556-021-00659-0. [DOI] [PubMed] [Google Scholar]
- Yu YS, Liu Y, An WS, Song JW, Zhang YF, Zhao XX STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. The Journal of Clinical Investigation. 2019;129(2):546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Hu ZP, Shi XL, Li XH, Zhan XM, Li XD, et al MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014;346(6216):1486–1492. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zeng WW, Chen ZJ MITAgating viral infection. Immunity. 2008;29(4):513–515. doi: 10.1016/j.immuni.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Zhang CG, Shang GJ, Gui X, Zhang XW, Bai XC, Chen ZJ Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019a;567(7748):394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu YT, Zhu YZ, Zhang Q, Guan HX, Liu SD, et al A non-canonical cGAS-STING-PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nature Cell Biology. 2022;24(5):766–782. doi: 10.1038/s41556-022-00894-z. [DOI] [PubMed] [Google Scholar]
- Zhang GG, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, et al Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proceedings of the National Academy of Sciences of the United States of America. 2016a;113(8):E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Liao BW, Xu ZS, Ran Y, Wang DP, Yang Y, et al USP44 positively regulates innate immune response to DNA viruses through deubiquitinating MITA. PLoS Pathogens. 2020a;16(1):e1008178. doi: 10.1371/journal.ppat.1008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hu MM, Wang YY, Shu HB TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. Journal of Biological Chemistry. 2012;287(34):28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Zhao J, Xu SM, Li JH, He SP, Zeng Y, et al Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host & Microbe. 2018;24(2):234–248.e5. doi: 10.1016/j.chom.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhang MX, Zhang Q, Zhu GF, Yuan L, Zhang DE, et al USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Research. 2016b;26(12):1302–1319. doi: 10.1038/cr.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MX, Cai Z, Zhang M, Wang XM, Wang YQ, Zhao F, et al USP20 promotes cellular antiviral responses via deconjugating K48-Linked ubiquitination of MITA. The Journal of Immunology. 2019b;202(8):2397–2406. doi: 10.4049/jimmunol.1801447. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Tang Z, An R, Ye LY, Zhong B USP29 maintains the stability of cGAS and promotes cellular antiviral responses and autoimmunity. Cell Research. 2020b;30(10):914–927. doi: 10.1038/s41422-020-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu JX, Du FH, Xu H, Sun LJ, Chen Z, et al The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Reports. 2014;6(3):421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XZ, Wu HX, Liu CG, Sun X, Zu SP, Tian J, et al The function of feline stimulator of interferon gene (STING) is evolutionarily conserved. Veterinary Immunology and Immunopathology. 2016c;169:54–62. doi: 10.1016/j.vetimm.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Zhao BY, Xu PB, Rowlett CM, Jing T, Shinde O, Lei YJ, et al The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature. 2020;587(7835):673–677. doi: 10.1038/s41586-020-2749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JN, Xiao RX, Zeng RQ, He ED, Zhang A Small molecules targeting cGAS-STING pathway for autoimmune disease. European Journal of Medicinal Chemistry. 2022;238:114480. doi: 10.1016/j.ejmech.2022.114480. [DOI] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao FC, et al The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zhong B, Zhang L, Lei CQ, Li Y, Mao AP, Yang Y, et al The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30(3):397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Zhong L, Hu MM, Bian LJ, Liu Y, Chen Q, Shu HB Phosphorylation of cGAS by CDK1 impairs self-DNA sensing in mitosis. Cell Discovery. 2020;6:26. doi: 10.1038/s41421-020-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang YF, Yang FH, Mao HQ, Chen Z, Zhang L Mitochondrial DNA leakage induces odontoblast inflammation via the cGAS-STING pathway. Cell Communication and Signaling. 2021a;19(1):58. doi: 10.1186/s12964-021-00738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Lin H, Wang SY, Wang S, Ran Y, Liu Y, et al The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host & Microbe. 2014;16(4):450–461. doi: 10.1016/j.chom.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, et al Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell. 2018;174(2):300–311.e11. doi: 10.1016/j.cell.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Whiteley AT, Kranzusch PJ Analysis of human cGAS activity and structure. Methods in Enzymology. 2019;625:13–40. doi: 10.1016/bs.mie.2019.04.012. [DOI] [PubMed] [Google Scholar]
- Zhou YL, He CX, Wang L, Ge BX Post-translational regulation of antiviral innate signaling. European Journal of Immunology. 2017;47(9):1414–1426. doi: 10.1002/eji.201746959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhang XY, Lei XB, Xiao X, Jiao T, Ma RY, et al Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduction and Targeted Therapy. 2021b;6(1):382. doi: 10.1038/s41392-021-00800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhou WL, Wang JX, Puc J, Ohgi KA, Erdjument-Bromage H, et al A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Molecular Cell. 2007;27(4):609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QY, Zhang YL, Wang L, Yao XY, Wu D, Cheng JJ, et al Inhibition of coronavirus infection by a synthetic STING agonist in primary human airway system. Antiviral Research. 2021;187:105015. doi: 10.1016/j.antiviral.2021.105015. [DOI] [PMC free article] [PubMed] [Google Scholar]