Abstract
Mosquitoes are known to harbor a large number of insect specific viruses (ISV) in addition to viruses of public health importance. These ISVs are highly species specific and are non-pathogenic to humans or domestic animals. However, there is a potential threat of these ISVs evolving into human pathogens by genome alterations. Some ISVs are known to modulate replication of pathogenic viruses by altering the susceptibility of vector mosquitoes to pathogenic viruses, thereby either inhibiting or enhancing transmission of the latter. In the present study, we report predominance of Phasi Charoen-like virus (PCLV, Family: Phenuviridae) contributing to >60% of the total reads in Aedes aegypti mosquitoes collected from Pune district of Maharashtra state using next generation sequencing based metagenomic analysis of viromes. Similar results were also obtained with mosquitoes from Assam, Tamil Nadu and Karnataka states of India. Comparison of Pune mosquito sequences with PCLV Rio (Brazil) isolate showed 98.90%, 99.027% and 98.88% homologies in the S, M and L segments respectively indicating less genetic heterogeneity of PCLV. The study also demonstrated occurrence of transovarial transmission as seen by detection of PCLV in eggs, larvae, pupae and male mosquitoes. Ae. aegypti mosquitoes collected from Pune also showed a large number of reads for viruses belonging to Baculoviridae, Rhabdoviridae, Genomoviridae and Bunyaviridae families. The role of PCLV in the replication of dengue and chikungunya virus is yet not clear. It warrants further studies to know the significance of PCLV and other ISVs on the replication and transmission of Ae. aegypti borne pathogenic viruses, especially in the absence of prophylactics or therapeutics.
Introduction
Emerging and re-emerging arthropod borne viruses continue to be a major threat to humanity as they not only inflict high morbidity and mortality but also devastate the economy of many countries especially in the tropical and subtropical countries [1–4]. During the last few decades, mosquito-borne viruses have emerged as a major public health concern, replacing malaria that had the dubious distinction as a major mosquito borne killer disease across the globe. Recent efforts on diagnosis and therapy have brought down malaria cases in most of the endemic countries, while therapeutics/vaccines for arboviral infections still remain scanty. The increased frequency and speed of human travel and commercial trade combined with global warming has resulted in geographic expansion of mosquito vectors and their pathogens from indigenous habitats to newer/naïve areas. The emerging /re-emerging viruses have the potential of inducing large fatal outbreaks involving the whole country or a continent in a short time span. The chikungunya virus (CHIKV) which re-emerged in Kenya in 2004, spread like a wild fire causing large-scale outbreaks with high morbidity in several countries of Asia, Europe, Oceania, the Caribbeans and the South and North Americas [1,2]. Similar is the case with West Nile virus (WNV), that caused massive outbreaks in North America, South America and Europe with high morbidity and mortality, since it was first detected in New York in 1999 [3,4].
Aedes mosquitoes, especially Aedes aegypti has garnered special attention due to its rapid geographical spread and public health importance. It has already established itself in the tropical and subtropical climate zones. The day biting urban mosquito is highly anthropophilic and is the principal vector of some of the important viruses, viz., dengue, chikungunya, yellow fever and Zika [5]. Dengue is endemic to almost all the tropical and subtropical countries with estimated 390 million cases annually [6]. In the absence of effective vaccines or therapeutics as well as mosquito control, dengue continues to be a major health concern today. The control of arboviral infections still largely relies on the vector management. Recent studies documenting reduced replication and transmission of dengue virus in Aedes mosquitoes infested with bacteria of Wolbachia species seems to be a promising novel biological control means [7,8]. Promising results have also been observed with certain insect specific viruses (ISVs) that alter the susceptibility of vector mosquitoes or cell lines derived from mosquitoes to human pathogenic viruses [9,10]. These studies have shown inhibition/ reduced replication of Japanese encephalitis, Murray Valley encephalitis and West Nile viruses in respective vector mosquitoes/ mosquito cells that were experimentally infected with ISVs. These studies suggest possibility of using ISVs as biocontrol agents since they cause homologous interference in the hosts by blocking the receptors [11]. However, ISVs are also regarded as the precursor to several human pathogenic viruses and therefore need special attention as they have the potential to emerge and cause human infections [12].
Studies conducted in China, Grenada and other countries have reported the prevalence of Phasi Charoen-like virus (PCLV) in Aedes aegypti populations, however, its potential to influence replication of viruses like dengue and chikungunya is not yet clear [13,14]. In the present study, we report predominance of PCLV in Aedes aegypti mosquitoes collected from Pune, Maharashtra, and three other states of India, i.e., Karnataka, Tamil Nadu and Assam.
Materials and methods
(i) Mosquito collection and processing
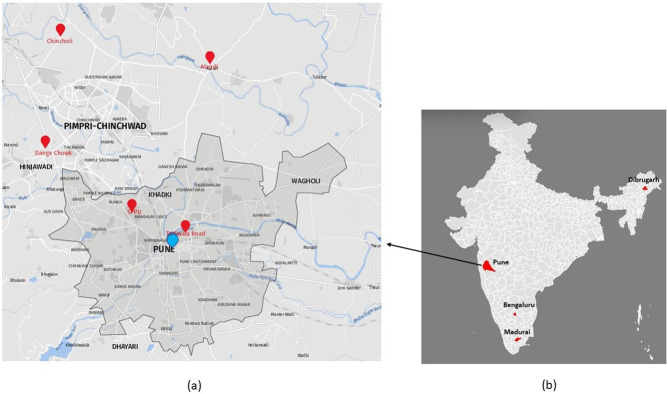
Adult Aedes aegypti mosquitoes were collected using hand held mouth aspirators, identified using entomological keys [15], pooled according to gender and locality and stored at -86°C until processing. Aedes larvae collected from breeding habitats were brought to ICMR-National Institute of Virology (NIV), Pune laboratory and reared until adults. The larvae reared adults were identified individually, pooled and used for the study. The mosquitoes were maintained at 28°C with 80–85% humidity and a 12:12 h photoperiod. Mosquitoes from Pune district of Maharashtra comprised collections from five different locations, viz., Dange Chowk, Savitribai Phule Pune University campus, Tadiwala Road, Alandi and Chincholi while that of Karnataka and Assam represented by mosquitoes from Bengaluru and Dibrugarh respectively. The Tamil Nadu mosquitoes represented the laboratory colony established at NIV from mosquito eggs received from ICMR-Vector Control Research Centre, Madurai, Tamil Nadu (Fig 1). Only female mosquitoes obtained from five locations of Pune were processed for the total virome analysis. For other locations, adult mosquitoes, eggs, larvae and pupae were processed for RT-PCR based detection of PCLV using S region specific primers (Table 1).
Fig 1. Geo-reference map of locations from where the mosquitoes were collected.
1a) The enlarged map of Pune Municipal Corporation (PMC), Pune City, prepared using software available at https://www.mapz.com (with due permission) shows two locations (Savitribai Phule Pune University area and Tadiwala road) while three locations on the outskirt of Pune city (Alandi, Chincholi and Dange chowk), 1b) Map of India prepared using software at https://www.gramener.com (free software) showing four locations from India, viz., Bengaluru, Dibrugarh, Madurai and Pune.
Table 1. Primers used in the study.
| Primer type | Primer | Primer sequence (5’ to 3’) | Reference |
|---|---|---|---|
| Random anchored primer | RT5 | CATCACATAGGCGTCCGCTGNNNNNN | Xiao et al. [16] |
| Random anchored primer | RT10 | CGACCCTCTTATCGTGACGGNNNNNN | Xiao et al. [16] |
| Random anchored primer | RT12 | GGTGGGCGTGTGAAATCGACNNNNNN | Xiao et al. [16] |
| Random anchored primer | IDT- K-8N | GACCATCTAGCGACCTCCACNNNNNNNN | Chrzastek et al. [17] |
| Anchor primer | P5 | CATCACATAGGCGTCCGCTG | Xiao et al. [16] |
| Anchor primer | P10 | CGACCCTCTTATCGTGACGG | Xiao et al. [16] |
| Anchor primer | P12 | GGTGGGCGTGTGAAATCGAC | Xiao et al. [16] |
| Anchor primer | IDT-K | GACCATCTAGCGACCTCCAC | Chrzastek et al. [17] |
| L segment- Forward | AGACAGCACAAGCAAATAAAGCAAG | Ramos-Nino et al. [13] | |
| L segment-Reverse | AAACATGCATTGTAAGGTTTTGTCG | Ramos-Nino et al. [13] | |
| M segment-Forward | AAAAGTAGGAATTGATGCTGTTGC | Ramos-Nino et al. [13] | |
| M segment-Reverse | CTTTGAGCACTTTTGTCTAATGGC | Ramos-Nino et al. [13] | |
| S segment Forward | CAGTTAAAGCATTTAATCGTATGATAA | Ramos-Nino et al. [13] | |
| S segment Reverse | TGGAAAATAAAAACAATAAAGCAATAC | Ramos-Nino et al. [13] |
(ii) Next generation sequencing based Aedes aegypti virome analysis using Oxford Nanopore technology
Mosquito homogenization and sample processing
Viral RNA was extracted from pools of 20–50 Aedes aegypti female mosquitoes. Briefly, the mosquitoes were surface sterilized using absolute ethanol for first wash followed by two consecutive washes with 70% ethanol and a final wash with sterile milliQ water/nuclease-free water. Surface sterilized mosquitoes were transferred to a sterile pre-chilled mortar and the frozen mosquito tissue was crushed in liquid nitrogen using a pestle until the mosquito tissue was completely homogenized as a fine powder. The homogenized tissue was suspended in 1X PBS (phosphate buffered saline) containing 5 mM MgCl2 and 1.4 mM Dithiothreitol (DTT, Invitrogen) and centrifuged at 17,000 g for 10 min at 4°C to pellet down cellular debris.
Viral RNA enrichment
Clear supernatants were collected and filtered twice through sterile 0.22 μM syringe filters to exclude bacterial and other larger microscopic entities. Filtrates were treated with 2 U/μl TURBO DNase (Invitrogen), 10 U/μl RNase A (Invitrogen) and incubated at 37°C for one hour to digest mosquito nucleic acids, bacterial and other genomes, essentially to enrich the enveloped/capsid-protected viruses.
Optimization of the workflow for NGS based virome analysis
To optimize the workflow for library preparation, lysates were prepared using pools of 50 Ae. aegypti female mosquitoes (from laboratory colony) and spiked with cell culture grown West Nile virus (WNV), Japanese encephalitis virus (JEV), dengue virus (DENV) (serotype two), Zika virus (ZIKV) and chikungunya virus (CHIKV) particles, equivalent to 104 plaque forming units each (to mimic realistic viral loads in mosquitoes) and processed further for library preparation. The primers for cDNA synthesis and amplification were selected based on broad metagenomic sensitivity for pathogen detection from previous reports [16–20]. A two-step strategy was optimized for cDNA synthesis using random anchored primers wherein first strand synthesis was done using reverse transcriptase while the second strand synthesis was done by using Klenow fragment. Four, out of fourteen, random anchored primers were selected for carrying out cDNA synthesis based on the properties, i.e., low self-annealing tendency, similar annealing temperatures and low tendency of primer dimer formation. Primer concentrations, reaction conditions and thermal cycling conditions were optimized to obtain ds-cDNA concentration equivalent to the input viral RNA template concentration. Sequence Independent Single Primer Amplification (SISPA) strategy was used for amplification of the cDNA fragments generated from viral RNA genomes.Library preparation was carried out using standard protocols provided by the Oxford Nanopore Technologies (ONT) for the Nanopore platform as described below using different primer sets (Table 1). The reaction conditions that yielded maximum sequence coverage (>90%) and depth for the spiked virus genomes were further used for processing the field collected mosquitoes from Pune.
Viral RNA extraction and library preparation
Viral RNA was extracted using QIAamp Viral RNA Mini Kit (Qiagen) as per the manufacturer’s instructions. Additional two washes with 80% and 70% ethanol were given on the column before sample elution to remove PCR inhibitors and to improve the quality of RNA. The eluted RNA was quantified using Qubit flourometer (Invitrogen).
Reverse transcription and double strand cDNA synthesis
RNA was subjected to first strand cDNA synthesis using four random anchored primers at a concentration of 20 picomoles (Table 1). For that, random anchored primers; one primer to each reaction, were added and incubated at 75°C for 5 min to allow denaturation of RNA secondary structures; and subjected to snap-cooling by placing on ice for 5 min to allow annealing of the primers to the template RNA. The first strand reaction final volume of 20 μl consisted of input RNA and primer, 1.0 μl SuperScript III Reverse Transcriptase (RT) (Invitrogen), 1.0 μl 0.1M DTT (Invitrogen), 1.0 μl (40U) RNase OUT (Invitrogen), 1.0 μl 25mM dNTPs and 4.0 μl 5 X first-strand buffer (Invitrogen). The reaction was carried out as follows: 25°C 10 min, 50°C 60 min, and 75°C 10 min. The first-strand cDNA synthesis reaction was followed by treatment with 250 U RNase H (NEB; New England Biologicals) to digest template RNA strand. Second strand cDNA synthesis was carried out using Klenow fragment (NEB). For that, the corresponding random anchored primer (Table 1) was added to the first-strand reaction and incubated at 75°C for 5 min, followed by snap-cooling on ice for 5 min. The final volume of 30 μl consisted of the first strand reaction, 10 picomoles of random anchored primer, 2.0 μl 10X Klenow buffer (NEB) and 200 U of Klenow fragment (NEB). The reaction was carried out at 37°C for 60 min followed by heat inactivation at 70°C for 10 min.
Sequence-Independent Single-Primer Amplification (SISPA)
The double stranded cDNA was subjected to SISPA, using anchor primers corresponding to the respective random anchored primers (Table 1). The 50 μl SISPA reaction consisted of 5.0 μl ds-cDNA, 2.5 μl anchor primer, 1.0 μl 25mM dNTPs, 0.5μl AmpliTaq Polymerase (Invitrogen), 10X PCR buffer (Invitrogen) and 1.25 μl 25mM MgCl2 (Invitrogen). The reaction was carried out at 95°C for 3 min, followed by 35 cycles at 95°C for 20 sec; 54°C for 45 sec; 72°C for 1 min and a final extension at 72°C for 7 min.
SISPA reaction was followed by a magnetic bead purification step using AMPure XP magnetic beads (Beckman-Coulter) to purify amplicons as per the manufacturer’s instructions. The amplicons were eluted in nuclease-free water and quantified using Qubit fluorometer. Only samples with quantities higher than 100 μg were taken forward for library preparation to ensure inclusion and representation of viral species from mosquito samples.
Library preparation and sequencing on Oxford Nanopore MinION Mk1B sequencer
DNA repair, blunt-end preparation and adapter ligation of amplicons were done as per the standard protocol for library preparation recommended by ONT using NEBNext FFPE DNA Repair Mix (M6630) and NEBNext Ultra II End repair / dA-tailing Module (E7546). The DNA sample was then subjected to clean-up with AMPure XP beads, eluted and quantified on Qubit fluorometer. Approx. 200–300 ng of amplified DNA of each sample (100 to 200 femtomoles) was ligated to sequencing adapters using SQK-LSK 109 kit (ONT) and barcoding was done using native barcoding EXP-NBD104 kit (ONT). Equal quantities of barcoded samples were pooled to 300ng for final sequencing and the remaining libraries were stored at -80˚C. Adapter ligated libraries were loaded onto the SpotON flow-cell (R.9.0.4) (MinION Mk1B system) and sequencing runs were performed for about 24 h using MinKNOW software (ONT).
Basecalling and data QC
The generated fast5 files were processed for basecalling using Guppy_basecaller v4.0.15 with high accuracy config file (dna_r9.4.1_450bps_hac.cfg). The reads in fastq format were demultiplexed (only if both the ends had the barcodes) using Guppy_barcoder v4.0.15. Only the reads with a minimum q-score of 7 were considered for further analysis. Porechop v0.2.4 was used for trimming all the adapter and barcode sequences. The resulting clean reads were used for further analysis.
Metagenomic classification
The reads were classified using Centrifuge v1.0.4 against a database of viral genomes. The genome specific reads were sorted and mapped against respective genomes using minimap2 and the coverage was calculated. The bcftools was used for assembling the mapped reads and generating the consensus genome sequence.
Confirmation of presence of PCLV with conventional RT-PCR and sequencing
Fresh viral RNA was extracted from Pune mosquito pools as described above. Forward and reverse pairs of primers as given in Table 1 were used for amplification of ~900–1000 nucleotide sequences from L, M and S segments of PCLV genome by RT-PCR using SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (Invitrogen). The amplicons were purified and both strands were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and analyzed on ABI 3130xl Genetic analyzer (Applied Biosystems, Foster City, USA). Mosquito samples collected from different states were processed similarly for viral RNA isolation and used for S gene amplification and sequencing.
Vertical transmission of PCLV
Eggs, larvae, pupae and adult mosquitoes reared from Ae. aegypti eggs from Madurai (Tamil Nadu) were processed to determine whether vertical transmission of the virus is occurring in the Aedes aegypti mosquitoes of the region. Generated amplicons were processed for sequencing and phylogenetic analysis.
Phylogenetic analysis
The obtained sequences were trimmed to remove primer sequences and aligned with various reference sequences downloaded from NCBI GenBank, using the ClustalW algorithm available through the Molecular Evolutionary Genetics Analysis (MEGA) 5 software [21]. Following this, the best fitting model for phylogenetic analysis was computed, and found to be the Tamura 3-Parameter model [22]. Maximum Likelihood phylogenetic trees were constructed using this model, taking uniform rates and 1000 bootstrap replicates.
Results
Optimization of library preparation for metagenome analysis
Lysates prepared from the laboratory colony of mosquitoes were spiked with cell culture grown WNV, JEV, dengue (serotype two), ZIKV and CHIKV particles and processed for viral RNA isolation, cDNA synthesis and library preparation using four different sets of primers. This was done in order to have the best conditions to carry out the metagenomic virome analysis of field mosquitoes. Library prepared with each of the P5 and P10, P12 and IDT-K primer set generated ~3 million reads. The trimmed reads were aligned onto the reference genomes of dengue, CHIKV, WNV, ZIKV, JEV and Aedes aegypti genome using minimap2, independently. All four primers, P5 and P10, P12 and IDT-K amplified the spiked viruses generating reads with the N50 of around 698 bases. Among the spiked viruses, dengue genome was recovered with maximum coverage 99.5%, while WNV, JEV and ZIKV genomes showed ~90% coverage with P5 and P10 primers. There were no reads representing chikungunya virus with both P5 and P10 primers. Though both P12 and IDT-K primers were able to successfully amplify CHIKV sequences, IDT-K primer (random octamer) showed >90% genome recovery. Pooling of libraries generated with primers P10 and IDK-K yielded sequence reads showing >90% coverage of genomes of all spiked viruses and hence it was decided to use both P10 and IDT-K primers for amplification in further experiments of virome analysis.
Metagenomic virome analysis of field caught Aedes aegypti mosquitoes
Field caught mosquitoes were processed for library preparation and sequencing using protocols optimized with virus spiking experiment. Each total virome sequencing run generated ~6.0 Gb data after 24 hours of the sequencing. After basecalling, demultiplexing, and trimming of adapters and primer sequences, average length of the reads was found to be ~300 bases (S1 Table). After removing the host Ae. aegypti sequences and reads <100 of length, the remaining clean reads were classified using BLASTn tool. The percentage of classified reads ranged between 3 to 53% for different runs after classification against a database containing human and viral genomes from RefSeq database (Table 2).
Table 2. Classified reads after classification against a database containing human and viral genomes.
| Sample | Classified reads | % classified reads | Chordate reads | Viral reads | PCLV reads | PCLV % in virome |
|---|---|---|---|---|---|---|
| RUN2_barcode01 | 18573 | 6.93 | 8343 | 10209 | 9685 | 94.87 |
| RUN2_barcode03 | 6408 | 2.88 | 3529 | 2877 | 2804 | 97.46 |
| RUN2_barcode04 | 522282 | 51.29 | 521008 | 1259 | 604 | 47.97 |
| RUN2_barcode05 | 141041 | 30.34 | 131983 | 8901 | 3013 | 33.85 |
| RUN2_barcode06 | 255523 | 30.02 | 242491 | 12906 | 8927 | 69.17 |
| RUN2_barcode07 | 322213 | 48.38 | 287419 | 34736 | 29867 | 85.98 |
| RUN2_barcode08 | 87601 | 14.72 | 47884 | 39541 | 33189 | 83.94 |
| RUN3_barcode01 | 340 | 38.12 | 336 | 4 | 3 | 75.00 |
| RUN3_barcode02 | 1021 | 39.93 | 1005 | 16 | 12 | 75.00 |
| RUN3_barcode03 | 1825 | 51.77 | 1755 | 70 | 22 | 31.43 |
| RUN3_barcode04 | 528 | 13.14 | 341 | 182 | 105 | 57.69 |
| RUN3_barcode05 | 258 | 12.60 | 179 | 79 | 44 | 55.70 |
| RUN4_barcode01 | 201193 | 11.72 | 121277 | 79145 | 44009 | 55.61 |
| RUN4_barcode02 | 154833 | 15.34 | 125205 | 29520 | 21630 | 73.27 |
| RUN4_barcode03 | 260004 | 34.92 | 257119 | 2825 | 1934 | 68.46 |
| RUN4_barcode04 | 132896 | 14.65 | 122883 | 9914 | 7331 | 73.95 |
| RUN4_barcode05 | 362075 | 29.01 | 354058 | 7826 | 2923 | 37.35 |
| RUN5_barcode01 | 325579 | 52.86 | 323486 | 2031 | 734 | 36.14 |
| RUN5_barcode02 | 91354 | 34.45 | 89156 | 2147 | 1059 | 49.32 |
| RUN5_barcode03 | 90009 | 28.16 | 83713 | 5674 | 2432 | 42.86 |
| RUN5_barcode04 | 20988 | 15.63 | 20272 | 696 | 9 | 1.29 |
| RUN5_barcode05 | 846 | 25.29 | 826 | 20 | 8 | 40.00 |
| RUN5_barcode06 | 1390 | 13.78 | 1205 | 184 | 8 | 4.35 |
The virus metagenomic analysis revealed abundance of Phasi Charoen-like virus (PCLV) belonging to Phenuviridae family of the order Bunyavirales in all the mosquito samples from Pune. PCLV constituted an average of 62.25% of the total mapped viral reads. On further mapping of these against the PCLV L, M and S genome segments to estimate the genome coverage, eleven out of seventeen processed mosquito pool samples showed more than 70% coverage of PCLV genome, while two samples showed more than 90% virus genome coverage (S2 Table). The metagenome sequences generated from Ae. aegypti mosquitoes from Pune are deposited in GenBank (S3 Table; Accession Nos. five locations and SRA IDs respectively). In addition to PCLV, reads of other viruses, viz., Choristoneura fumiferana granulovirus (ChfuGV) (Baculoviridae), Piry vesiculovirus (PIRYV) (Rhabdoviridae), Human Gemykibivirus 2 (HuGkV-2) (Genomoviridae), Shamonda virus (Bunyaviridae) were also detected in the samples (Table 3). Since the reads for these viruses were comparatively few and genome coverage and depth were comparatively low, these sequences were not analyzed further.
Table 3. Species identified with minimum 50 reads.
| Species | Taxon ID | Maximum reads classified | RUN2_NB01 | RUN2_NB03 | RUN2_NB04 | RUN2_NB05 | RUN2_NB06 | RUN2_NB07 | RUN2_NB08 | RUN4_NB01 | RUN4_NB02 | RUN4_NB03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | 9606 | 521008 | 8343 | 3529 | 521008 | 131983 | 242491 | 287419 | 47884 | 121277 | 125205 | 257119 |
| Phasi Charoen-like phasivirus | 1980610 | 44009 | 9685 | 8804 | 6604 | 3013 | 8927 | 29867 | 33189 | 44009 | 21630 | 1934 |
| Choristoneura fumiferana granulovirus | 56947 | 21118 | 166 | 12 | 14 | 188 | 203 | 2239 | 2620 | 21118 | 5774 | 296 |
| Piry vesiculovirus | 1972575 | 12405 | 209 | 7 | 569 | 5519 | 3622 | 2440 | 2057 | 12405 | 1475 | 512 |
| Human associated gemykibivirus 2 | 2004957 | 1011 | 1 | 0 | 2 | 77 | 1 | 82 | 1011 | |||
| Shamonda orthobunyavirus | 159150 | 727 | 4 | 0 | 5 | 3 | 11 | 38 | 174 | 47 | 48 | |
| Cotesia congregata bracovirus | 39640 | 112 | 0 | 9 | 15 | 91 | 112 | |||||
| Lactobacillus virus LP65 | 298338 | 73 | 13 | 0 | 1 | 2 | 1 | 7 | 13 | 18 | 2 | |
| Prochlorococcus phage P-SSM2 | 268746 | 73 | 0 | 0 | 10 | |||||||
| Mycobacterium virus Goose | 1211282 | 57 | 0 | 0 | 57 | |||||||
| Gokushovirinae Bog1183_53 |
1655646 | 54 | 0 | 0 |
Confirmation of the presence of PCLV in Aedes aegypti mosquitoes by conventional RT-PCR and sequence analysis
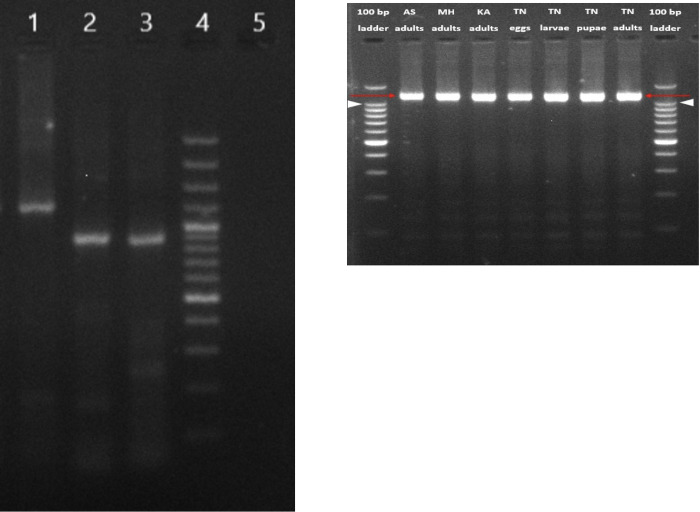
Viral RNA isolated from Ae. aegypti female mosquitoes collected from one location in Pune was processed for amplification of partial L, M, and complete S segment of PCLV genome to validate the NGS results. All the three segments could be successfully amplified (Figs 2A and S1). Sequencing of amplicons obtained from these segments using Sanger’s method confirmed these as PCLV sequences and validated the results obtained with NGS based metagenomic analysis. Sequence comparisons with the reference PCLV Rio isolate showed 98.90%, 99.027% and 98.88% homologies in the S, M and L segments respectively. These findings confirmed findings of NGS based metagenome analysis of viruses suggesting that PCLV is present abundantly in wild, field caught Ae. aegypti mosquitoes from Pune district, Maharashtra.
Fig 2.
a: Detection of PCLV S, M and L segments from Pune adult female mosquito pools using RT-PCR. Products were analysed on 2% agarose gel, lanes- L1: Complete S segment, L2: Partial M segment, L3: Partial L segment, L4: 100 bp DNA ladder, L5: Negative control. b: Detection of PCLV S segment from mosquito samples from different locations from India using RT-PCR. Products were analyzed on 2% agarose gel, lanes- L1: 100 bp DNA ladder (Invitrogen), L2: Assam (AS) adult mosquito pool, L3: Pune, Maharashtra (MH) adult mosquito pool, L4: Karnataka (KA) adult mosquito pool, L5: Tamil Nadu (TN) mosquito eggs, L6: Tamil Nadu (TN) mosquito larvae, L7: Tamil Nadu (TN) mosquito pupae, L8: Tamil Nadu (TN) mosquito adults, L9: 100 bp DNA ladder.
Detection of PCLV in Aedes aegypti mosquitoes collected from other states of India
PCLV ‘S’ segment was amplified from Ae. aegypti mosquito samples collected from Karnataka, Assam and Tamil Nadu states of India (Figs 2B and S2). Presence of PCLV in Ae. aegypti mosquitoes in Karnataka has already been reported recently [23]. PCLV S segment sequences from different locations of India were compared to reference PCLV S segment sequences across the world. The lowest percent nucleotide identity observed was 99.90% between S segment sequences generated from Ae. aegypti mosquitoes from Assam, India, and adult mosquitoes from Rio (Brazil). The highest homology observed was 99.98% between sequences from pupae of Tamil Nadu mosquitoes and adults from Kenya (Table 4). Ae. aegypti mosquitoes from the four states showed >99.95% homology of PCLV ‘S’ gene sequences. Overall results indicate that PCLV is a highly conserved virus and viral strains circulating in Ae. aegypti mosquitoes across the world have very high genetic similarity suggesting a common ancestor for these strains (Fig 2B).
Table 4. Percent Nucleotide identities of PCLV S segment sequences from various life cycle stages of Ae. aegypti mosquitoes collected from different parts of India compared to PCLV sequences across the world.
| Origin |
NC_038263.1 Rio_Brazil |
LC498493.1 Ghana |
MH237597.1 Australia |
MN053771.1 Guadeloupe |
MT361067.1 Kenya |
KM001087.1 Thailand |
MN109951.1 Grenada |
KU936055.1 UK |
MH310081.1 USA |
MF614134.1 China |
|---|---|---|---|---|---|---|---|---|---|---|
| IND (TN) (eggs) | 98.939 | 99.961 | 99.957 | 99.975 | 99.983 | 99.981 | 99.970 | 99.959 | 99.959 | 99.959 |
|
IND (TN)
(larvae) |
98.942 | 99.952 | 99.952 | 99.966 | 99.974 | 99.972 | 99.961 | 99.954 | 99.954 | 99.954 |
| IND (TN) (pupae) | 98.958 | 99.961 | 99.957 | 99.979 | 99.986 | 99.981 | 99.981 | 99.959 | 99.959 | 99.963 |
|
IND (TN)
(female) |
98.934 | 99.961 | 99.956 | 99.979 | 99.983 | 99.977 | 99.977 | 99.957 | 99.957 | 99.959 |
| IND (MH) | 98.901 | 99.950 | 99.941 | 99.956 | 99.967 | 99.959 | 99.952 | 99.943 | 99.943 | 99.946 |
| IND (AS) | 98.900 | 99.957 | 99.944 | 99.965 | 99.979 | 99.970 | 99.963 | 99.946 | 99.946 | 99.948 |
| IND (KN) | 98.901 | 99.948 | 99.939 | 99.957 | 99.968 | 99.961 | 99.954 | 99.941 | 99.941 | 99.941 |
Transovrial transmission of PCLV in Aedes aegypti
To determine whether transovarial transmission of PCLV occurs in Ae. aegypti mosquitoes, a colony was established from Ae. aegypti eggs obtained from Madurai (Tamil Nadu state) and larvae, pupae, adults and eggs from the colony were processed for S gene amplification. PCLV was detected in all the stages of Ae. aegypti indicating occurrence of transovarial transmission (vertical transmission) of the virus in Ae. aegypti. Nucleotide identities as compared to reference sequence were: eggs 99.984%, larvae 99.981, pupae 99.983% and adults 99.983% (Fig 3).
Fig 3. Phylogenetic tree for PCLV S segment sequences.
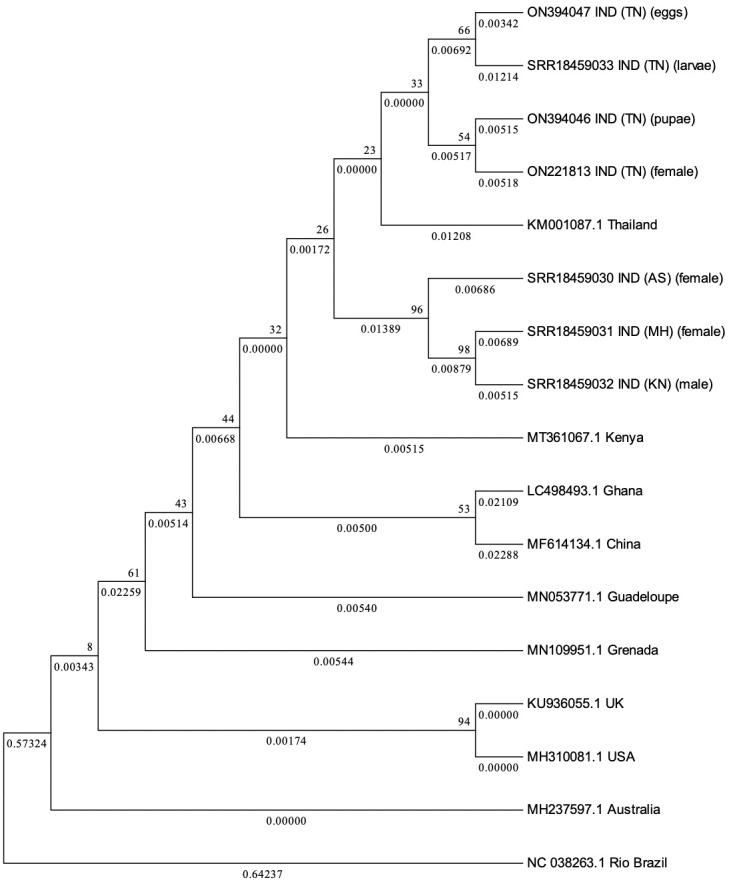
Phylogenetic tree constructed via Maximum Likelihood (ML) method, using Tamura 3-Parameter and 1000 Bootstrap replicates for PCLV S segment sequences obtained from Aedes aegypti mosquitoes from different states of India, along with reference sequences from across the world.
Discussion
Metagenomic analysis using Next generation sequencing platforms has become a powerful tool for analysis of genomes of organisms. A study by Shi et al [24] has reported presence of >1500 novel RNA viruses in invertebrates. Subsequently, several investigators across the globe have used the tool and identified novel viral reads in important disease vectors viz., Aedes aegypti, Culex quinquefasciatus, etc. The surge in dengue across the world in the recent decades has stimulated renewed research interest in Ae. aegypti mosquitoes as control of dengue still depends on vector management. It is speculated that vectorial capacity of mosquitoes can be influenced by the presence of endosymbiotic microbes by altering the susceptibility of the mosquitoes to pathogenic viruses [25]. Studies have also shown that endosymbiont organisms in mosquitoes inhibit or reduce the replication of pathogenic viruses and could be used as biological control agents [10,13,24]. Studies using Wolbachia, an enosymbiont bacterium in mosquitoes, have also shown that it can inhibit the replication of many eukaryotic viruses [7,26]. Aedes aegypti mosquitoes, which lack the natural colonization of Wolbachia have shown resistance to dengue and chikungunya viruses when experimentally infected with Wolbachia [27]. Culex quinquifasciatus mosquitoes harboring Wolbachia have also shown resistance to West Nile virus upon infection [28]. Since no licensed vaccine is available for dengue today, these studies may evolve in the development of suitable control agents to limit the growth of the virus in Aedes aegypti mosquito.
In the present study, we analysed virome of Ae. aegypti mosquitoes collected from different parts of India, i.e., Pune district of Maharashtra, Bengaluru (Karnataka), Dibrugarh (Assam) and Madurai (Tamil Nadu). We observed abundance of PCLV sequences in the mosquito samples tested from all the states. In complete virome analysis of field caught Ae. aegypti mosquitoes from Pune region of Maharashtra, PCLV constituted >60% reads of the total mapped viral reads. The rest constituted by an array of viruses, viz., members of virus families Baculoviridae, Rhabdoviridae, Genomoviridae, Bunyaviridae etc. Among these, Piry virus (PIRYV) (Family: Rhabdoviridae, genus: Vesiculovirus) and Human Gemykibivirus 2 (HuGkV-2) (family: Genomoviridae) (Table 3) are suspected to be human pathogens hence need further attention [29]. Piry virus (PIRYV) (genus Vesiculovirus) is similar to Chandipura virus [30] that shows very high mortality in infected children. Serosurveys from different regions of Brazil have shown 4–17.7% seropositivity against PIRYV suggestive of human exposure to the virus. It was noted that accidental human exposure to PIRYV induces symptoms such as fever, headache, weakness, myalgia and arthralgia indicating that the virus has potential to cause human disease [31]. Importantly, experimental PIRYV infections in adult Swiss albino mice induces neuropathological damage and behavioral changes [32] suggesting severe manifestations upon infection. Hence PIRYV warrants further studies to assess potential threat to humans. Though clinical impact of HuGkV-2 is currently not known, detection of HuGkV-2 DNA in human blood samples from Brazilian Amazon also draws attention towards this virus [33]. Reads detected for another virus requiring attention was, Shamonda virus belonging to the Simbu group of the genus Orthobunyavirus of the family Peribunyaviridae. Orthobunyaviruses are transmitted between mammalian hosts by arthropod vectors. Their tripartite genomes facilitate reassortment of genomic segments. Detection of Shamonda virus in Ae aegypti adult mosquitoes collected from different locations from Pune indicates that these mosquitoes are persistently infected with the virus. PCLV also belongs to the same family. With rapid global movement and favorable climatic changes for emergence of new viruses, it would be essential to monitor Peribunyavirus activity in mosquitoes. It would be worthwhile to isolate these mosquito viruses and do further studies to understand whether these viruses have ability to interfere in the growth of each other and of the viruses of medical importance. The metagenomic virome analysis did not show presence of any viruses of medical importance such as DENV, CHIKV and ZIKV in the field caught mosquitoes, possibly since there were no current outbreaks. We used RNA metagenomic approach using the Oxford Nanopore sequencing platform to generate nearly complete genome sequence of Phasi Charoen-like virus infecting field-caught Ae. aegypti female mosquitoes in Pune, Maharashtra, India. Analysis of Ae. aegypti mosquitoes from three other states of India also revealed that this virus is abundantly present in these mosquitoes. Furthermore, presence of the virus at all developmental stages of the mosquito indicated vertical transmission of PCLV. Since PCLV is insect specific virus and there is no intermediate vertebrate host that can amplify the virus, it appears that transovarial transmission is the only way to maintain the virus.
Importantly, comparative sequence analysis showed >99% homology between Indian isolates as well as in isolates of other countries, suggesting high genetic stability of the virus. Presence of PCLV in natural populations of Ae. aegypti without significant changes in viral genomes suggests its possible endosymbiotic or commensal relationship with the mosquito without harming the fitness of its host.
Detection of PCLV in Ae. aegypti mosquitoes has already been reported from Brazil, Grenada, China, Thailand, USA etc [13,14,24,25,34]. However, the implication of PCLV in Ae. aegypti mosquitoes in the inhibition of pathogenic viruses such as dengue or chikungunya is not clearly known. Entomological investigations during dengue/chikungunya outbreaks in different parts of India have shown transmission by Ae. aegypti mosquitoes despite harboring high density of PCLV suggestive of its negative role in blocking the virus transmission (NIV unpublished data). This is in agreement with in vitro studies by Fredericks et al [5] where they have shown persistently infected cells harboring PCLV, did not show any impact on culturing and growth of other arboviruses. Being a member of the family Peribunyaviridae, which comprises several viruses of public health importance; the role of PCLV in interfering with replication of bunyaviruses is important and warrants investigation. Presence of ISVs belonging to family Flaviviridae, viz., Bagaza virus, Palm Creek virus etc., have shown inhibition of JEV, WNV and Murray Valley encephalitis virus in their respective vectors [9,10]. Experimental studies have shown the phenomenon of superinfection exclusion where cultures/mosquitoes infected with one flavivirus, reduced replication of another flavivirus upon subsequent infection [35].
Not much is still known about the role played by PCLV and other viruses in Aedes aegypti mosquitoes in the replication and transmission of human pathogenic viruses, viz., dengue, chikungunya, Zika or Yellow fever. Further research into these aspects may come up with the potential of using the mosquito specific viruses in managing/impacting Ae. aegypti borne pathogenic viruses especially in the absence of prophylactics or therapeutics.
Supporting information
Products were analysed on 2% agarose gel, lanes- L1: Complete S segment, L2: Partial M segment, L3: Partial L segment, L4: 100 bp DNA ladder, L5: Negative control.
(TIFF)
Products were analyzed on 2% agarose gel, lanes- L1: 100 bp DNA ladder (Invitrogen), L2: Assam (AS) adult mosquito pool, L3: Pune, Maharashtra (MH) adult mosquito pool, L4: Karnataka (KA) adult mosquito pool, L5: Tamil Nadu (TN) mosquito eggs, L6: Tamil Nadu (TN) mosquito larvae, L7: Tamil Nadu (TN) mosquito pupae, L8: Tamil Nadu (TN) mosquito adults, L9: 100 bp DNA ladder.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(PDF)
Acknowledgments
The authors wish to thank Professor Priya Abraham, the Director, ICMR- NIV, for her keen interest in the project and for the constant support. The authors also thank Dr Ashwani Kumar and Dr R Paramsivan from ICMR-Vector Control Research Centre, Puducherry and Dr Siraj Khan, Regional Medical Research Centre Dibrugarh, Assam for providing mosquito eggs for the study.
Data Availability
All relevant data are within the article and its supporting information files.
Funding Statement
Indian Council of Medical Research, New Delhi, India funded the project as an extramural project granted to ABS, KSL and SC. Grant No. 6/9-7 (2019-ECD-II). The funders had no role in study design, data collection and analyis, decision to publish or preparation of the manuscript.
References
- 1.Petersen LR, Powers AM. Chikungunya: epidemiology. F1000Res. 2016. Jan 19;5:F1000 Faculty Rev-82. doi: 10.12688/f1000research.7171.1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim SM, Dutta SK and Martina BEE. Defining efficacy of chikungunya virus candidate vaccines: different endpoints derived from the virus—cytokine—ferritin (VCF) model. Front. Virol. 2021; 1:693439. doi: 10.3389/fviro.2021.693439 [DOI] [Google Scholar]
- 3.Danforth ME, Fischer M, Snyder RE, Lindsey NP, Martin SW, Kramer VL. Characterizing areas with increased burden of West Nile Virus disease in California, 2009–2018. Vector Borne Zoonotic Dis. 2021. Aug;21(8):620–627. doi: 10.1089/vbz.2021.0014 Epub 2021 Jun 2. ; PMCID: PMC8380797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronca SE, Ruff JC, Murray KO. A 20-year historical review of West Nile virus since its initial emergence in North America: Has West Nile virus become a neglected tropical disease? PLoS Negl Trop Dis. 2021; 15(5): e0009190. doi: 10.1371/journal.pntd.0009190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredericks AC, Russell TA, Wallace LE, Davidson AD, Fernandez-Sesma A, Maringer K Aedes aegypti (Aag2)-derived clonal mosquito cell lines reveal the effects of pre-existing persistent infection with the insect-specific bunyavirus Phasi Charoen-like virus on arbovirus replication. PLoS Negl Trop Dis 2019;13(11): e0007346. doi: 10.1371/journal.pntd.0007346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013. Apr 25;496(7446):504–7. doi: 10.1038/nature12060 Epub 2013 Apr 7. ; PMCID: PMC3651993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson K N. The Impact of Wolbachia on virus infection in mosquitoes. Viruses 2015; 7: 5705–5717; doi: 10.3390/v7112903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Utarini A, Indriani C, Ahmad RA, Tantowijoyo W, Arguni E, Ansari MR, et al. AWED Study Group. Efficacy of Wolbachia-Infected mosquito deployments for the control of dengue. N Engl J Med. 2021. Jun 10;384 (23):2177–2186. doi: 10.1056/NEJMoa2030243 ; PMCID: PMC8103655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudeep AB, Bondre VP, George R, Ghodke YS, Aher RV, Gokhale MD. Bagaza virus inhibits Japanese encephalitis and West Nile virus replication in Culex tritaeniorhynchus & Cx. quinquefasciatus mosquitoes. Indian J Med Res 2015; 142: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobson-Peters J, Yam AWY, Lu JWF, Setoh YX, May FJ, et al. A new insect-specific flavivirus from Northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013; 8(2): e56534. doi: 10.1371/journal.pone.0056534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Öhlund P., Lundén H. & Blomström AL. Insect-specific virus evolution and potential effects on vector competence. Virus Genes 55, 127–137 (2019). doi: 10.1007/s11262-018-01629-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elrefaey AM, Abdelnabi R, Rosales Rosas AL, Wang L, Basu S, Delang L. Understanding the mechanisms underlying host restriction of insect-specific viruses. Viruses. 2020. Aug 31;12(9):964. doi: 10.3390/v12090964 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos-Nino ME, Fitzpatrick DM, Tighe S, Eckstrom KM, Hattaway LM, Hsueh AN, et al. High prevalence of Phasi Charoen-like virus from wild-caught Aedes aegypti in Grenada, W.I. as revealed by metagenomic analysis. PLoS One. 2020. Jan 31;15(1):e0227998. doi: 10.1371/journal.pone.0227998 ; PMCID: PMC6993974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Huang S, Jin T, Lin P, Huang Y, Wu C, et al. Discovery and high prevalence of Phasi Charoen-like virus in field-captured Aedes aegypti in South China. Virology. 2018. Oct; 523:35–40. doi: 10.1016/j.virol.2018.07.021 [DOI] [PubMed] [Google Scholar]
- 15.Barraud PJ. The fauna of British India, including Ceylon and Burma. Diptera.Vol V. Family Culicidae. Tribes Megarhinini and Culicini. London: Taylor and Francis; 1934. [Google Scholar]
- 16.Xiao P, Han J, Zhang Y, Li C, Guo X, Wen S, et al. Metagenomic Analysis of Flaviviridae in mosquito viromes isolated from Yunnan Province in China reveals genes from dengue and Zika viruses. Front Cell Infect Microbiol. 2018;8:359. doi: 10.3389/fcimb.2018.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chrzastek K, Lee DH, Smith D, Sharma P, Suarez DL, Pantin-Jackwood M, et al. Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology. 2017. Sep; 509: 159–166. doi: 10.1016/j.virol.2017.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Li Z, Yang F, Zheng J, Feng Y, Guo H, et al. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS ONE 2013, 8:e61950. doi: 10.1371/journal.pone.0061950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Q, Wan L, Li X, Cao X, Zhou Y, Song C. Metagenomic evidence reveals denitrifying community diversity rather than abundance drives nitrate removal in storm water bio filters amended with different organic and inorganic electron donors. Chemosphere. 2020. Oct;257:127269. doi: 10.1016/j.chemosphere.2020.127269 [DOI] [PubMed] [Google Scholar]
- 20.Hameed M, Liu K, Anwar MN, Wahaab A, Li C, Di D, et al. A viral metagenomic analysis reveals rich viral abundance and diversity in mosquitoes from pig farms. Transbound Emerg Dis. 2020. Jan;67(1):328–343. doi: 10.1111/tbed.13355 [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011. Oct;28(10):2731–9. doi: 10.1093/molbev/msr121 Epub 2011 May 4. ; PMCID: PMC3203626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992. Jul;9(4):678–87. doi: 10.1093/oxfordjournals.molbev.a040752 . [DOI] [PubMed] [Google Scholar]
- 23.Munivenkatappa A, Nyayanit DA, Yadav PD, Rangappa M, Patil S, Majumdar T, et al. Identification of Phasi Charoen-Like Phasivirus in field collected Aedes aegypti from Karnataka State, India. Vector Borne Zoonotic Dis. 2021;21(11):900–909. doi: 10.1089/vbz.2021.0011 Epub 2021 Sep 14. [DOI] [PubMed] [Google Scholar]
- 24.Shi C, Beller L, Deboutte W, Yinda KC, Delang L, Vega-Ru´a A, et al. Stable distinct core eukaryotic viromes in different mosquito species from Guadeloupe, using single mosquito viral metagenomics. Microbiome. 2019; doi: 10.1186/s40168-019-0734-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandler JA, Thongsripong P, Green A, Kittayapong P, Wilcox BA, Schroth GP, et al. Metagenomic shotgun sequencing of a Bunyavirus in wild-caught Aedes aegypti from Thailand informs the evolutionary and genomic history of the Phleboviruses. Virology. 2014; doi: 10.1016/j.virol.2014.06.036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker T, Moriera LA. Can Wolbachia be used to control malaria? Mem Inst Oswaldo Cruz, Rio de Janeiro, 2011; Vol. 106 (Suppl. I): 212–217. doi: 10.1590/s0074-02762011000900026 [DOI] [PubMed] [Google Scholar]
- 27.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139: 1268–1278. [DOI] [PubMed] [Google Scholar]
- 28.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE. 2010; 5: e11977. doi: 10.1371/journal.pone.0011977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupovic M, Ghabrial SA, Jiang D, Varsani A. Genomoviridae: a new family of widespread single-stranded DNA viruses. Arch Virol. 2016. Sep;161(9):2633–43. doi: 10.1007/s00705-016-2943-3 Epub 2016 Jun 24. . [DOI] [PubMed] [Google Scholar]
- 30.Cherian SS, Gunjikar RS, Banerjee A, Kumar S, Arankalle VA. Whole genomes of Chandipura virus isolates and comparative analysis with other rhabdoviruses. PLoS One. 2012;7(1):e30315. doi: 10.1371/journal.pone.0030315 Epub 2012 Jan 17. ; PMCID: PMC3260278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tavares-Neto J, Travassos da Rosa AP, Ataide M, Morais-Souza H, Vasconcelos P, Travassos da Rosa J (1990) Frequency of neutralizing antibodies to the vesiculovirus Piry, in blood donors of Uberaba, Minas Gerais, Brazil. Rev Inst Med Trop Sao Paulo 32:211–214. [PubMed] [Google Scholar]
- 32.Zucherato VS, Giovanetti M, Costa LOA, Krause LMF, Alves DCC, Moreira RMA, et al. Molecular identification of the emerging Human Gemykibivirus-2 (HuGkV-2) among Brazilian blood donors. Transfus Apher Sci. 2022. Jul 26:103516. doi: 10.1016/j.transci.2022.103516 Epub ahead of print. . [DOI] [PubMed] [Google Scholar]
- 33.de Sousa AA, Reis R, Bento-Torres J, Trevia N, Lins NA, Passos A, et al. (2011) Influence of enriched environment on viral encephalitis outcomes: behavioral and neuropathological changes in albino Swiss mice. PLoS One 6:e15597. doi: 10.1371/journal.pone.0015597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunha MDP, Ioshino RS, Costa-da-Silva AL, Petersen V, Capurro ML, Zanotto PMA. A Metagenomic Approach Identified a Novel Phasi Charoen-Like virus co-infecting a chikungunya virus-Infected Aedes aegypti mosquito in Brazil. Microbiol Resour Announc. 2020. Jul 30;9(31):e01572–19. doi: 10.1128/MRA.01572-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goenaga S, Goenaga J, Boaglio ER, Enria DA, Levis SDC. Superinfection exclusion studies using West Nile virus and Culex flavivirus strains from Argentina. Mem Inst Oswaldo Cruz. 2020. Jun 3;115:e200012. doi: 10.1590/0074-02760200012 [DOI] [PMC free article] [PubMed] [Google Scholar]