Abstract
Immunofluorescence is used in numerous research areas including eye research to detect specific antigens in cells and tissues. One limitation is that fluorescent signal can fade, causing detection problems if data recording was not completed in a timely manner or if additional data acquisition is required. The ability to repeat immunostaining for the same antigen after initial fluorescence has faded may require time-consuming and potentially damaging steps to remove primary antibodies. Our studies assessed whether immunofluorescence could be reapplied to previously labeled retinal ganglion cells (RGCs). To examine whether immunostaining of Brn3a, a commonly used RGC marker, could be repeated in retinas with previously faded immunostaining, retinal whole mounts were labeled with anti-Brn3a primary antibodies and green fluorescent secondary antibodies, then allowed to fade over time. Faded retinas were restained with anti-Brn3a antibody followed by secondary antibody, or with secondary antibody alone. Results show restaining with anti-Brn3a primary antibody followed by Alexa-fluor green secondary antibody is effective for RGC detection. Repeat RGC labeling improved the clarity of staining compared with original staining prior to fading, with significant reduction in the percentage of blurry/out of focus fluorescent cells (6 vs 26%); whereas, repeat application of secondary antibody alone was not effective. Preflattening retinas under a coverslip prior to initial Brn3a staining also increased the clarity of staining, and facilitated significantly more accurate automated counting of RGCs. Findings suggest Brn3a antigen remains accessible for repeat immunofluorescence labeling after original staining fades. Staining retinas after flattening tissue may enhance the clarity of staining and accuracy of automated RGC counting. Repeat immunofluorescence staining, without the need to strip off prior bound antibodies, may be useful in other tissues as well and warrants future examination.
Keywords: Immunofluorescence, Brn3a staining, retinal ganglion cells
1. Introduction
Immunohistochemistry is an integral part of cell research used to highlight cells in many different fields. In eye research, immunohistochemistry is used to identify and quantify specific populations of cells, such as retinal ganglion cells (RGCs), the cells that comprise the optic nerve, by immunostaining using a cell-type specific antibody. Brn3a is a Pit-Oct-Unc family transcription factor activated in post-mitotic RGCs (Latchman, 1999), and anti-Brn3a immunofluorescence is a commonly used method of identifying RGCs in retinal tissues (Gramlich et al., 2020; Guo et al., 2021; Khan et al., 2021; Nadal-Nicolás et al., 2009; Ross et al., 2021). One potential limitation of immunofluorescence is that the staining can fade with time and from light exposure (Johnson, et al., 1982), even with various strategies including limiting light exposure and use of mounting reagents that reduce fading (Valnes and Brandtzaeg, 1985) employed to prevent loss of signal. For studies using Brn3a immunofluorescence to quantify RGC survival, investigators typically take pictures before staining fades, but if this is delayed, the ability to measure RGC numbers may be lost. Similarly, if additional data is required, such as quantification of larger areas of the retina, faded staining may prevent repeat analysis of previously stained tissues.
When initial immunofluorescence staining fades, it may be possible to re-stain markers of interest, but it is unclear how much antigen remains accessible for repeat staining without added steps designed to remove the original primary antibody. With additional steps, reports suggest repeat antibody staining may be effective. Multiplexing, staining with multiple antibodies and repeated rounds of staining, has shown that immunofluorescence staining can be performed multiple times on a single tissue section (Bolognesi et al., 2017); however, removal of primary antibodies by disulfide cleavage was required, and repeat staining was performed with different antibodies as opposed to restaining twice with the same antibody. Multiplex immunohistochemical staining can also be performed after de-staining of hematoxylin & eosin (Hinton et al., 2019). Another study showed that repeated staining with different concentrations of single primary antibody leads to improved staining at lower concentrations than required with a single application of antibody (Smith, 2016), suggesting repeat staining might only be possible if antigen is not already saturated in the initial staining procedure. For Brn3a staining, the potential ability to restain retinal tissue is unclear. In this study, restained Brn3a slides were observed and counted in comparison with faded slides and originally stained slides. Results show that repeat staining, without the need to remove previously bound antibodies, is feasible and effective and could provide a simple approach for vision research investigators to obtain data that otherwise may have been lost.
2. Material and Methods
2.1. Mice
C57BL/6J mice were obtained from the Jackson Laboratory and raised in a 12 hour light/dark cycle. Animals were housed at the University of Pennsylvania. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania and were performed in compliance with the Association for Research in Vision and Ophthalmology statement for the Use of Animals in Ophthalmic and Vision Research as well as with institutional and federal regulations.
2.2. Initial and Preflattened Brn3a Staining
Immunostaining was performed as in prior studies (Khan et al., 2021; Ross et al., 2021). Prior to euthanasia, mice were anesthetized with 5% isoflurane and eyes were removed. Eyes were fixed in 4% paraformaldehyde for 1 hour at room temperature, then washed in phosphate buffered saline (PBS) 5 times for 5 minutes at room temperature. The retina of each eye was then isolated and prepared as a whole mount, washed three times in PBS for five minutes at room temperature, then permeabilized in 0.5% Triton X-100 in PBS by freezing at −70°C for 15 minutes. Additional retinas were prepared as whole mounts, coverslipped in mounting media (Fluoromount-G; Southern Biotech, Birmingham, AL) to induce increased flattening of tissue, and stored at 4°C for one week prior to removing the coverslips and permeabilizing the tissue before adding primary antibody. Retinas were then incubated overnight at 4°C in a humidified chamber with rabbit anti-Brn3a antibody (Synaptic Systems, Goettingen, Germany) diluted 1:2000 in blocking buffer (2% Triton X-100 and 2% bovine serum albumin). The next day, retinas were washed three times in PBS for five minutes each wash at room temperature. After washing, they were incubated in a humidified chamber for 1 hour at room temperature with Alexa Fluor 488 anti-rabbit secondary antibody (Thermo Fisher Scientific, Waltham, MA) diluted 1:500 in blocking buffer, then washed five more times in PBS for five minutes each wash at room temperature. Retinas were mounted with their vitreous side up on glass slides with Fluoromount-G mounting medium.
2.3. Brn3a Restaining after Fading
Slides were stored at 4 °C for 3–4 months and exposed to light to allow fluorescent signal to fade. As slides were visualized by fluorescence microscopy, photobleaching could be a contributing factor to signal fading. After fading, pictures were taken of the tissue to document the decrease in RGC immunostaining signal. Coverslips were removed using a razor blade taking care not to disturb underlying retina. The tissues were then washed 3 times in PBS for 5 minutes at room temperature, and the Brn3a staining procedure was repeated using the exact same procedure for both primary and secondary antibody staining as detailed for the initial staining. Alternatively, a separate group of retinas were not restained with anti-Brn3a antibody, but were incubated for 1 hour at room temperature in Alexa Fluor 488 anti-rabbit secondary antibody at a dilution of 1:500 in blocking buffer, then washed five times in PBS for five minutes each at room temperature and mounted with Fluoromount-G mounting medium.
2.4. Assessment of RGC staining
Brn3a-positive RGCs were photographed in a set of representative areas as in prior studies (Khan et al., 2021; Ross et al., 2021) taken by fluorescent microscopy (Nikon, Melville, NY) at a magnification of 40x in 12 standard fields: one, three and five sixths of the retinal radius from the center of the retina, in each retinal quadrant covering a total area of 0.407 mm2/retina. Photographs were taken using the autoexposure setting in the Nikon NIS-Elements AR 4.60.00 software. The pictures were analyzed using Image-Pro Plus 5.0 (Media Cybernetics, Silver Spring, MD) and RGCs were counted by an investigator masked to the experimental conditions. Counts were converted to RGCs/mm2 for comparisons between groups. To evaluate the clarity of staining, cells were counted in identical areas of each retina, one third of the retinal radius from the center. The quality of RGC staining was assessed by counting the number of Brn3a-positive cells that were judged to be blurry (out of focus) in each photograph, as assessed by a single investigator masked to the experimental conditions. The quality was determined by counting the percentage of blurry cells out of the total number of stained cells present.
2.5. Comparison of accuracy of human and automated counting of RGCs
Representative images were selected to highlight the level of clarity of Brn3a labeled cells measured in originally stained, restained, and preflattened retinas. Four representative images, taken from 4 separate retinas, were selected from each group, and brightness of all images was set to 70 and contrast to 150 using ImageJ software (NIH, Bethesda, MD). Each selected representative image was rotated clockwise by 180 degrees and also mirrored along the vertical midline using Adobe Photoshop, thus generating 3 copies of each image containing the same cells but in different orientations. All images (3 copies of each × 4 images/group × 3 staining protocols = 36 total images) were sorted into random order without allowing two inverted images of the same cells to be placed next to each other. The number of Brn3a positive cells were then counted by eight individuals masked to the staining conditions (one counter was excluded after they recognized that there were duplicate/mirrored images included). All counting was performed on ImageJ. In addition, automated counting was performed using the RGC Batch plugin in Fiji/ImageJ (Simple RGC), an open-source image processing software installed as described by Cross et al. (2021) after images were grayscaled by conversion from RGC color to 16-bit. Parameters were determined by visual inspection, with morphology channel = 1, cell diameter (px) = 30–200, local threshold radius = 200, Gaussian blur sigma = 1. Data output was received on Microsoft Excel.
2.6. Statistics
The number of Brn3a stained cells per high powered field counted in original stained retinas and retinas restained after fading was compared by student’s T-test. The percentage of Brn3a stained cells observed to be out of focus in original stained, preflattened, and restained retinas were compared by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons testing. The percentages of Brn3a-positive cells over- or under-counted (absolute value) by automated counting versus human counting in original stained, preflattened, and restained retinas were compared by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons testing. All comparisons were performed using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). A difference with a p-value less than 0.05 was considered statistically significant.
3. Results
3.1. Repeat Brn3a staining labels as many RGCs as original staining.
Mouse retinas were immunostained with anti-Brn3a antibodies and RGCs were photographed to document initial staining (Fig 1A). Slides were then stored for several weeks and periodically exposed to both ambient light and repeat fluorescence microscopy to induce fading of fluorescent signal, resulting in loss of identifiable cells (Fig 1B). Faded slides restained with primary anti-Brn3a antibody showed numerous RGCs (Fig 1C) similar to the original stained retina, whereas slides that were restained with the secondary antibody alone (Fig 1D) did not appear visibly different from the faded staining where little or no cells were visible. The number of RGCs per area was similar in retinas after original staining as compared with restaining after fading (Fig 1E).
Figure 1.
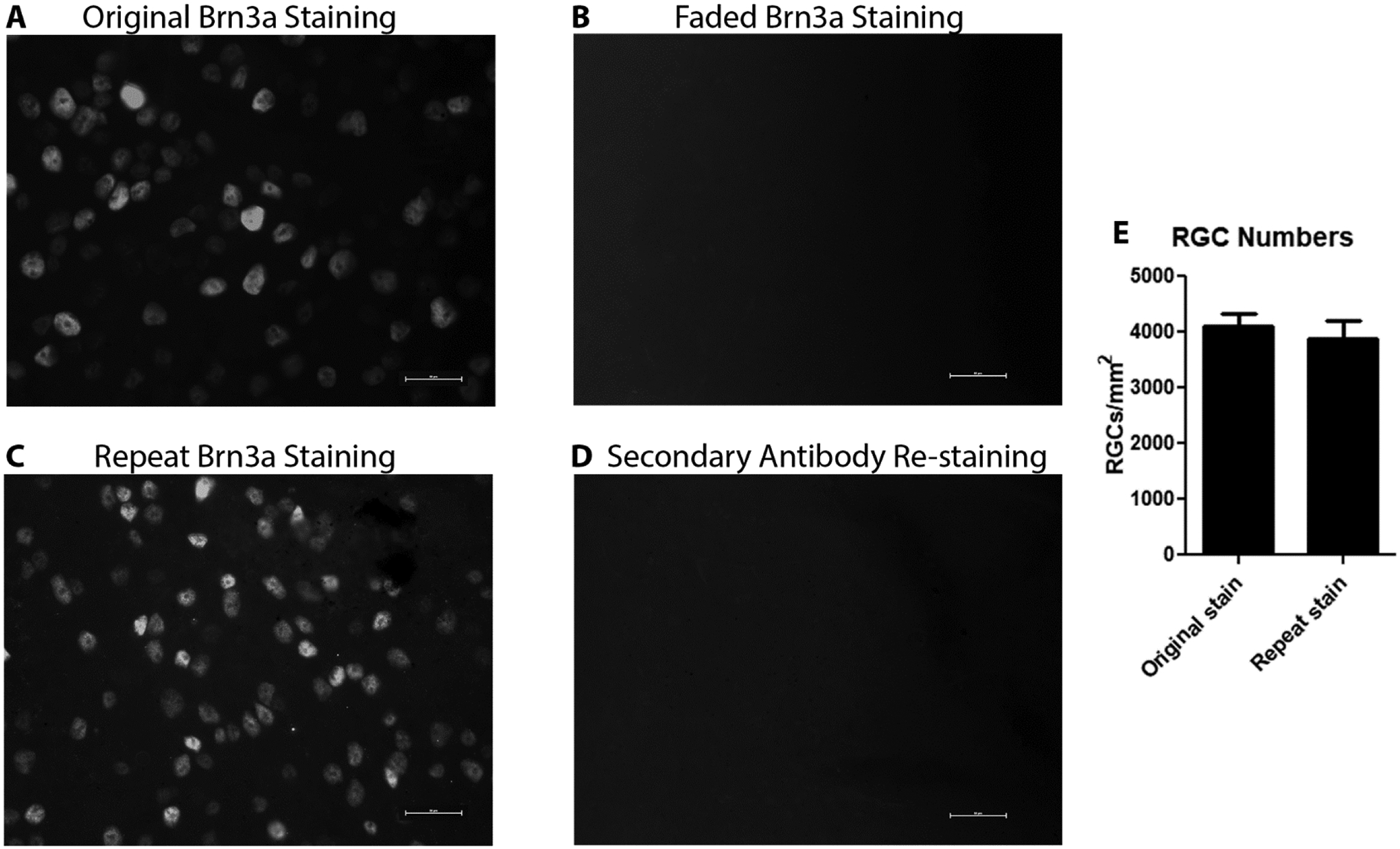
Representative photographs of retinas after original Brn3a staining, faded Brn3a staining, repeat Brn3a staining, and attempted secondary antibody repeat staining. Original Brn3a staining (A) shows numerous antigen positive cells. Faded staining (B) has no clearly defined cells. The restained retina (C) shows many Brn3a-postive cells, similar to the original stained slides. The secondary antibody only re-stained retina (D) is similar to the faded staining picture with no detectable signal. There is no significant difference in the average number of Brn3a-positive cells counted in standardized retinal fields between original stained and restained retinas (N = 6 retinas/group; p = 0.5858). Scale bars = 25 microns.
3.2. Restaining after fading or staining after preflattening improves clarity of Brn3a-positive cells.
The clarity of Brn3a immunofluorescence was assessed by a masked investigator to identify cells that were stained but were not in clear focus in retinal images (Fig 2A–C). When the original staining was compared to the restained slides quantitatively, the restained slides had significantly fewer blurry cells (Fig 2D), with 6.0±1.5% of cells judged to be blurry (N = 6 retinas) as compared with 25.9±5.1% of cells judged to be out of focus in retinas following initial Brn3a staining (N = 6; p < 0.01). A separate group of retinas flattened for one week on slides under coverslips prior to initial Brn3a staining also had significantly fewer out of focus cells (4.2±1.2%; N = 6) than retinas stained initially without added preflattening (p < 0.001).
Figure 2.
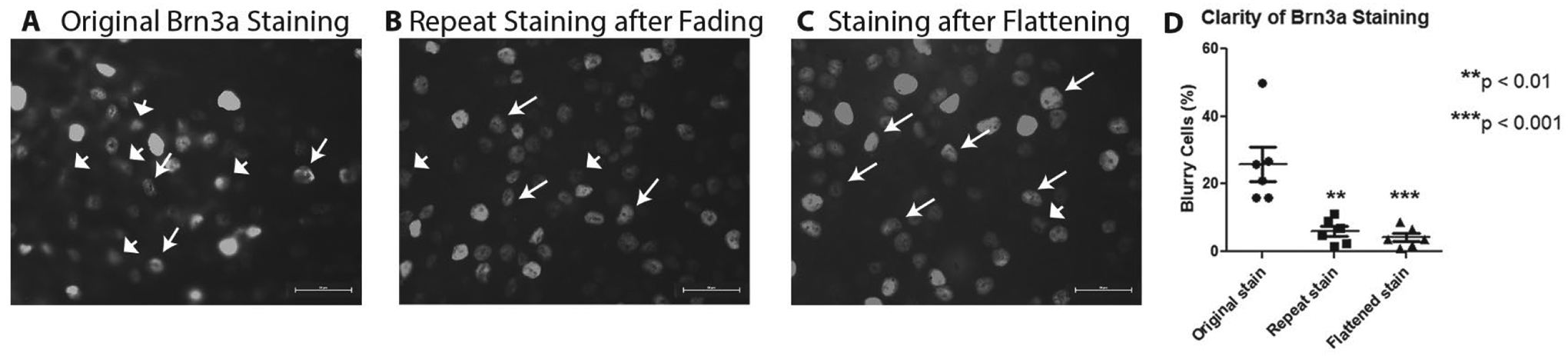
Representative photo of original Brn3a (A) staining, which has many cells both clear and out of focus; repeated Brn3a staining (B), which has many cells, most of which are clearly defined; and a retina pre-flattened under a coverslip for one week prior to original Brn3a staining (C), which also shows numerous cells in clear focus. (D) Comparison of percentages of blurry cells from original staining (N = 6 retinas), repeat staining (N = 6), and original staining after preflattening (N = 6) shows that significantly fewer RGCs are out of focus in re-stained (**p < 0.01) and pre-flattened (***p < 0.001) retinas as compared with original Brn3a staining. Scale bars = 25 microns.
3.3. Improved clarity of Brn3a staining by preflattening/restaining increases accuracy of automated RGC counting as compared with manual counting.
Four representative images of Brn3a-positive cells from each group of original stained, restained, and preflattened retinas were selected, then copied and rotated to include 3 different orientations of each image (Fig 3). The number of Brn3a-positive cells counted in each image by 7 different masked investigators, and by the Simple RGC (Cross et al., 2021) automated counting program, were used to assess consistency of cell counting. Both human and automated counting showed highly consistent counts from each counter, with an average standard error of the mean between the 3 counts of each copied image as just ±3 or less averaged across all human counters, and just ±1 or less for the Simple RGC program (Fig 3B). However, automated counting with Simple RGC demonstrated significantly more deviation from manually counted RGC numbers quantified by masked investigators in original stained retinas as compared with re-stained or preflattened retinas (Fig 3C; p < 0.001), as Simple RGC counted an average of 50% more Brn3a-postive cells than human counters in original stained retinas, but only over- or under-counted by less than 20% in restained and preflattened retinas.
Figure 3.
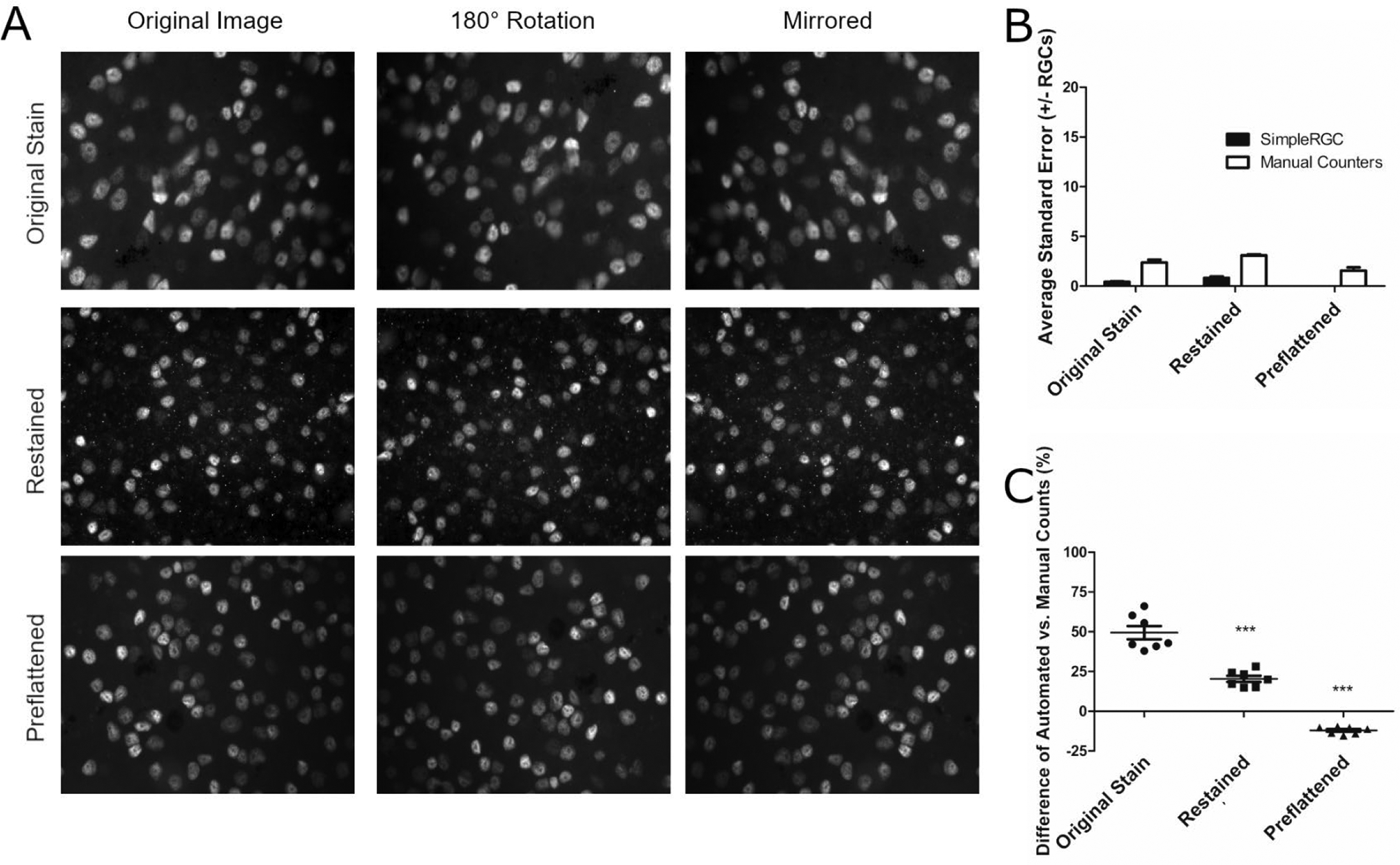
Preflattening retinas before staining improves concordance between human and automated cell counts. (A) Representative images of one original, one restained and one preflattened retina after staining for Brn3a showing respective image orientation manipulations: rotation 180 degrees clockwise and mirroring across the vertical meridian. Images were taken at 40x magnification and mixed in random order with images from other retinas before being counted by masked individuals. One of four images from each staining group is shown. (B) The standard error of the number of Brn3a-positive cells counted by each counter (human or automated) was calculated for each set of 3 duplicate/inverted images as shown in A. Seven masked human counters varied in their individual counts of three duplicate images (N = 4 sets of 3 images/group) by no more than ±3 cells regardless of whether retinas were originally stained, restained, or preflattened before staining. Simple RGC automated counting was also highly consistent with no more than ±1 cells between counts of 3 copies of each image regardless of whether retinas were originally stained, restained, or preflattened before staining (N = 4 sets of 3 images/group). (C) Differences in automated vs. manual counters are displayed as the percent difference between the automated Simple RGC count and each individual human counter (N = 7). Percent difference was calculated by subtracting automated means from the mean of each human counter and dividing by the human mean. The percent difference between automated and manual counts in original stained retinal images was significantly higher (***p<0.001) than the percent differences between automated and manual counts in restained or preflattened retinal images.
4. Discussion
Results show that when staining fades over time, RGCs can be re-stained using anti-Brn3a antibodies without a need to remove the original antibodies, thus avoiding time consuming and potentially damaging steps to strip off antibodies used in initial staining (Bolognesi et al., 2017). Of note, repeat staining was just as efficient as original staining, labeling an equivalent number of RGCs, albeit by indirect measurement of representative retinal fields. Direct assessment of efficiency of re-staining is limited by the inability to re-image the exact same fields. Nonetheless, results showing efficient repeat staining suggest that either the original primary antibody application did not saturate all available Brn3a epitopes, or the initial antibodies detached or degraded over time resulting in a sufficient amount of accessible Brn3a antigen for repeat antibody application to bind the full Brna3a-positive RGC population.
Surprisingly, repeat Brn3a staining led to improved quality of staining with fewer cells out of focus in retinal images. It is not clear why re-stained retinas had fewer cells out of focus, but it is likely that the tissues flattened over time on slides underneath the coverslips following their initial staining, bringing cells into a single focal plane better captured on imaging. Improved clarity of staining observed in retinas that were pre-flattened under coverslips prior to initial Brn3a staining further supports this possible explanation, as illustrated in Figure 4.
Figure 4.
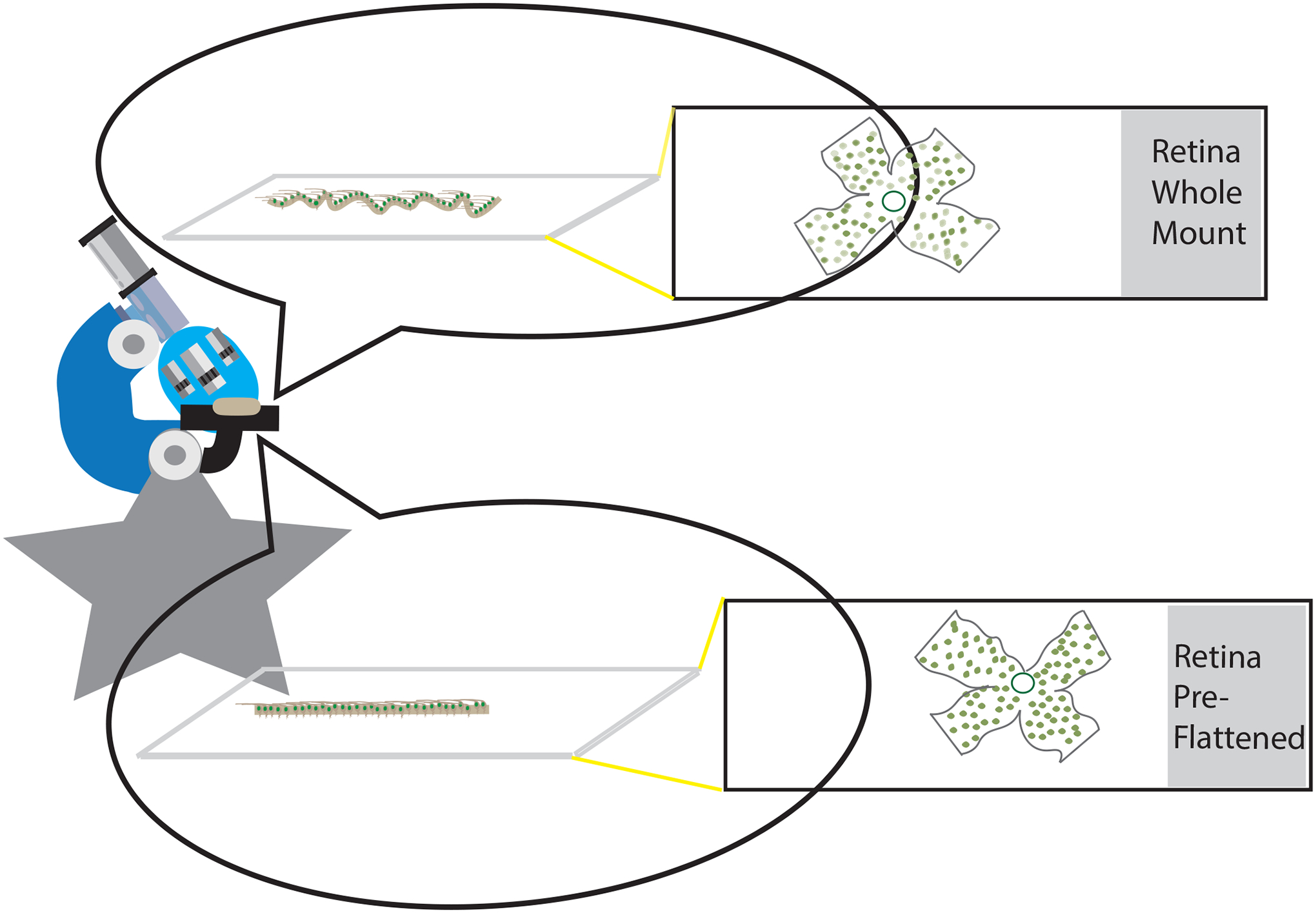
Illustration shows potential explanation for observed improvement in clarity of Brn3a staining after retinas have flattened over time under a coverslip prior to staining or restaining. Dissected retinas are flat-mounted on slides but tissue retains some thickness and microscopic folds/undulations (top) that can flatten further over time under a coverslip (bottom).
Interestingly, results show that masked, human investigators manually counting Brn3a-postivie cells and automated counting using the Simple RGC (Cross et al., 2021) program are both remarkably internally consistent in identifying cells, regardless of how clear the labeled cells appear in captured images. This is supported by data showing masked human counters varied by only ±3 cells when counting three rotated copies of the same images not only from clear images of restained and preflattened retinas, but also from selected original stained retinal images containing blurry/out of focus cells. Automated counting was also highly consistent with variations of only ±1 cell between copied images even in the original stained retinas containing blurry/out of focus cells. However, results do suggest that the threshold used by automated counting to identify cells can differ significantly from human counters when a notable subset of cells are blurry/out focus, as automated counts on average were 50% higher than counts from each of 8 masked investigators when counting original Brn3a-stained retinas. Significantly less deviation between automated and manual counts was found in restained or preflattened retinas. Thus, preflattening retinas prior to Brn3a staining might be useful if investigators plan to use automated counting to quantify RGC numbers.
5. Conclusion
Together, data and results here suggest a useful method to prevent lost data in RGC studies if Brn3a staining has faded prior to completing data acquisition. Results suggest preflattening retinas may lead to improved clarity of Brn3a staining and improved agreement between automated and manual counts of Brn3a-positive cells in the retina. Further investigation is warranted to determine whether this simple strategy is applicable to other immunohistochemical stains and tissues, as a number of factors including differences in antigenicity and stability of target proteins being probed could affect the outcome of the re-staining process
Highlights.
Repeat RGC staining is simple and highly effect
The quality of Brn3a staining is improved after tissue flattening
Repeat Brn3a staining is a useful method to prevent lost data in retinal studies, with further investigation warranted to determine whether this simple strategy is applicable to other immunohistochemical stains
Acknowledgements:
Funding provided by NIH Grants EY019014 and EY030163; Research to Prevent Blindness; the F. M. Kirby Foundation; and the RWJ Harold Amos Faculty Development Award. Authors thank Gordon Hua, Sage Cho, Jason Wu, Justin Guo, Bradford Fauntleroy, Adrian Corbey, Tiffany Atmadja, Nuala O’Neill, and Alex Kwok for their work as single-blind counters of Brn3a-positive cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors declare no conflicts of interest related to this work.
References
- Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, Cattoretti G. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J Histochem Cytochem. 65, 431–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross T, Navarange R, Son J-H, Burr W, Singh A, Zhang K. Rusu M, Gkoutzis K. Osborne A, Nieuwenhuis B. Simple RGC: ImageJ Plugins for Counting Retinal Ganglion Cells and Determining the Transduction Efficiency of Viral Vectors in RLatchman DS. POU family transcription factors in the nervous system. J Cell Physiol. 179, 126–133 (1999). [DOI] [PubMed] [Google Scholar]
- Gramlich OW, Brown AJ, Godwin CR, Chimenti MS, Boland LK, Ankrum JA, Kardon RH. Systemic Mesenchymal Stem Cell Treatment Mitigates Structural and Functional Retinal Ganglion Cell Degeneration in a Mouse Model of Multiple Sclerosis. Transl Vis Sci Technol. 9, 16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mehrabian Z, Johnson MA, Miller NR, Henderson AD, Hamlyn J, Bernstein SL. Biomarkers of lesion severity in a rodent model of nonarteritic anterior ischemic optic neuropathy (rNAION). PLoS One. 16, e0243186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton JP, Dvorak K, Roberts E, French WJ, Grubbs JC, Cress AE, Tiwari HA, Nagle RB. A Method to Reuse Archived H&E Stained Histology Slides for a Multiplex Protein Biomarker Analysis. Methods Protoc. 2, 86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Davidson RS, McNamee KC, Russell G, Goodwin D, Holborow EJ. Fading of immunofluorescence during microscopy: a study of the phenomenon and its remedy. J Immunol Methods. 55, 231–42 (1982). [DOI] [PubMed] [Google Scholar]
- Khan RS, Ross AG, Aravand P, Dine K, Selzer EB, Shindler KS. RGC and Vision Loss From Traumatic Optic Neuropathy Induced by Repetitive Closed Head Trauma Is Dependent on Timing and Force of Impact. Transl Vis Sci Technol. 10, 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Nicolás FM, Jiménez-López M, Sobrado-Calvo P, Nieto-López L, Cánovas-Martínez I, Salinas-Navarro M, Vidal-Sanz M, Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest Ophthalmol Vis Sci. 50, 3860–3868 (2009). [DOI] [PubMed] [Google Scholar]
- Ross AG, McDougald DS, Khan RS, Duong TT, Dine KE, Aravand P, Bennett J, Chavali VR, Shindler KS. Rescue of retinal ganglion cells in optic nerve injury using cell-selective AAV mediated delivery of SIRT1. Gene Ther. 28, 256–264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AA. Repeated immunostaining of the same tissue section using alkaline phosphatase as a reporter. Biotech Histochem. 91, 396–400 (2016). [DOI] [PubMed] [Google Scholar]
- Valnes K, Brandtzaeg P. Retardation of immunofluorescence fading during microscopy. J Histochem Cytochem. 33, 755–61 (1985). [DOI] [PubMed] [Google Scholar]


