Abstract
Telomere elongation is coupled with genome replication, raising the question of the repair of short telomeres in post-mitotic cells. We investigated the fate of a telomere-repeat capped end that mimics a single short telomere in quiescent fission yeast cells. We show that telomerase is able to elongate this single short telomere during quiescence despite the binding of Ku to the proto-telomere. While Taz1 and Rap1 repress telomerase in vegetative cells, both shelterin proteins are required for efficient telomere extension in quiescent cells, underscoring a distinct mode of telomerase control. We further show that Rad3ATR and Tel1ATM are redundantly required for telomere elongation in quiescence through the phosphorylation of Ccq1 and that Rif1 and its associated-PP1 phosphatases negatively regulate telomerase activity by opposing Ccq1 phosphorylation. The distinct mode of telomerase regulation in quiescent fission yeast cells may be relevant to that in human stem and progenitor cells.
INTRODUCTION
Terminal DNA of linear chromosomes consists of long tracts of G/C-rich short tandem repeats that end with a single-stranded 3′ overhang. These repeats are protected by several proteins constituting the shelterin complex (1). The resulting nucleoprotein structure called telomere preserves genome integrity by ensuring complete replication of terminal DNA, and establishing the spatial organization of the genome within the nucleus (2–4). At each round of DNA replication, telomeres shorten because of the incomplete replication of the linear DNA molecules by the conventional DNA polymerases (5). To circumvent this telomeric loss, shelterin proteins recruit telomerase that elongates the 3′ overhang by the addition of telomeric repeats (1,6,7). In human, during mid-embryogenesis, transcription of TERT, the catalytic subunit of telomerase, is shut down in most somatic cells, which leads to telomere shortening, and ultimately to replicative senescence (8). In contrast, TERT either remains expressed in adult stem cells, including intestines, skin and hematopoietic stem cells, or is transiently activated in certain proliferating stem-like cells. Upon differentiation, this transient expression extinguishes (9,10).
Although telomere attrition is prominent in proliferative tissues, telomere shortening has been also observed in terminally differentiated non-cycling somatic cells of brain regions or skeletal muscle (11,12), suggesting causes that are independent of cell divisions. Indeed, nucleoside base alteration by oxidative stress or other DNA damages may alter the binding of telomeric protein, trigger DNA repair, and accelerate telomere shortening (13–16). These observations raise the question of how telomeres are maintained in quiescence (in either post-mitotic or stem cells), especially given that telomerase activity is limited to S phase in most organisms studied (17–21). Indeed, evidences are mounting that the processing of telomeric DNA ends requires the passage of the replication fork (18,22,23). The capacity of stem cells to enter and exit quiescence is imperative for tissue homeostasis and responses to life-threatening challenges (24). Cancer stem cells can also oscillate between states of quiescence and proliferation. When in quiescence, they are resistant to conventional chemotherapy treatments, which explains frequent tumor relapse (25). They display shorter telomeres compared to normal stem cells that may influence their ability to exit quiescence (8). The foregoing observations highlight the importance of the understanding of how telomeres are maintained in quiescence and how their length impact the transition between quiescent and proliferative states.
Fission yeast Schizosaccharomyces pombe is an attractive model organism to study telomere biology since it has a shelterin-like complex that shares structural and functional similarities with the mammalian shelterin (26). In details, Taz1 associates to the double-stranded telomeric DNA and recruits Rif1 and Rap1. Pot1 binds to the 3′-overhang and is bridged to the Taz1 protein through a network of protein-protein interactions including Tpz1 and Poz1 (27,28). While Rif1, Taz1, Rap1 and Poz1 negatively regulate telomere lengthening, the complex Pot1-Tpz1-Ccq1 is important for telomere capping and telomerase recruitment (26,29). Indeed, the phosphorylation of Ccq1 protein, on Thr93 by both Rad3ATR and Tel1ATM kinases, triggers telomerase recruitment (30,31).
In addition, fission yeast cells can be easily maintained in quiescence by nitrogen starvation (G0), a simple nutritional change that yields reproducible results (32–34). Lately, we investigated the stability of telomeres in fission yeast cells that were maintained in quiescence (35). In cells lacking telomerase, we discovered that eroded telomeres were highly rearranged during quiescence, and that these rearrangements prevented proper re-entry into proliferative phase. More recently, we further demonstrated that nuclear envelop attachment of telomere limited these rearrangements (36). However, we did not address the capacity of these eroded ends to be repaired by telomerase in non-dividing cells.
To tackle this issue, we investigated the fate of a telomere-repeat capped end mimicking a single short telomere in quiescent fission yeast cells. Strikingly, we observed that telomerase is recruited and is active outside of S-phase in quiescent cells as reflected by its ability to elongate the short telomere. We reported that the Ku complex also binds to and protects the short telomere and regulates telomerase action. Unexpectedly, we found that Taz1 and Rap1 are both required for short telomere extension, in contrast to what was observed in vegetative cells in which the lack of Taz1 or and Rap1 results in elongated telomeres (37,38). Accordingly, the absence of Taz1 and Rap1 altered the recruitment of Ccq1 to the short telomere. Furthermore, both Rad3ATR and Tel1ATM were redundantly required for telomere elongation by telomerase through a process dependent on Ccq1 phosphorylation. Finally, we uncovered that Rif1 participates in the regulation of telomerase by antagonizing Ccq1 phosphorylation through its PP1 phosphatase activity. These results provide support for an unanticipated mode of telomerase action in quiescent fission yeast cells that contrasts in many aspects with telomerase control in cycling cells.
MATERIALS AND METHODS
Strains and growth conditions
The fission yeast strains used in this study are prototrophic and listed in Supplementary Table S1. Telomerase was deleted by substituting the ter1 gene with kanamycin cassette by one-step homologous insertion using FwTer1-5’UTR and RevTer1-3’UTR primers from genomic DNA. For G0 experiments, strains were grown in Edinburgh minimal medium with sodium glutamate substituted for ammonium chloride (PMG) and 100 μg/ml hygromycin B (InvitroGen) at 32°C (or at 25°C for stn1-226 thermosensitive mutant) to a density of 6 × 106 cells/ml, washed three times in minimal medium deprived from nitrogen (MM-N), and resuspended in MM-N at a density of 2 × 106 cells/ml at 32°C, reaching after two rounds of cell division 8 × 106 cells/ml. This density of cell bodies is stable for several weeks and was used as a source of G0 cells.
Telomere length analysis by Southern blotting
Genomic DNA was prepared from 2 × 108 cells and digested with ScaI-HF restriction enzyme (New England Biolabs). The digested DNA was resolved on 1.5% agarose gel and blotted onto a Hybond-XL membrane (GE Healthcare), as previously described (39). After transfer, the membrane was cross-linked with UV and hybridized with ura4 and abo1 probes made with primers listed in Supplementary Table S2 (40). 32P labeling of DNA probes was performed by random priming using Klenow fragment lacking exonuclease activity (New England Biolabs), in presence of [α-32P]dCTP (Perkin Elmer) and hybridizations were performed in Church buffer at 60°C for ura4 and abo1 probes (40). Radioactive signals were detected using a Biorad molecular imager FX. The telomere length was determined using the BioRad software.
Telomere PCR and sequencing
Telomere PCR was performed as previously described (40). Briefly, 200 ng of genomic DNA was suspended in a total of 5 μl of deionized water and heated at 95°C for 5 min. Tailing reaction was performed in a final volume of 10 μl with final concentrations of 1 mM dCTP, 15 units of Terminal Deoxynucleotide Transferase (Promega) and 1X Terminal Transferase Buffer (100 mM cacodylate buffer, 1 mM CoCl2, 0.1 mM DTT (pH 6.8)). Reaction mix was incubated at 37°C for 30 min, then at 70°C for 10 min to stop the reaction. The poly-dC-tailed DNA was then PCR amplified using AccuPrime™ GC-Rich DNA Polymerase (Invitrogen) with a primer specific for the ura4 region and a primer complementary to the poly-dC-tail listed in Supplementary Table S2. PCR program is: 3′ at 94°C, 45 cycles of 94°C 30’’; 55°C 30’’;72°C 30’’ followed by a final extension step of 5′ at 72°C. The PCR products were either purified by gel extraction (QIAquick Gel Extraction Kit) or using spin columns (QIAquick PCR purification Kit) and cloned into the pCRBLunt plasmid and sequenced using M13F primer.
ChIP
2 × 109, 4 × 108 or 7 × 108 cells were crosslinked with 1% of Formaldehyde (Sigma Aldrich) at room temperature for 15 min for Est1-V5, Ccq1-flag Ku-myc ChIP and Taz1-GFP, respectively. Crosslinking was quenched with 125 mM Glycine for 5 min at room temperature and cells were washed twice with ice-cold TBS. Cell pellets were then resuspended in Lysis Buffer Low Sodium (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-Deoxycholate, 2 mM of PMSF, protease inhibitors) and mechanically lysed with glass beads using 7 cycles of 20 s at speed 6.5. in fast-prep machine. Whole cell extracts were sonicated for 10 min (30 s ON, 30 s OFF) using the Diagenode Bioruptor. Sonicated lysates were cleared by centrifugation and standardized by nanodrop. Solubilized chromatin was prepared at equal DNA content before adding 2 μl of anti-V5 (R96025, Invitrogen), 2 μl of anti-myc (9E10 sc-40, Santa Cruz), 2 μl of anti-flag (M2, Sigma Aldrich) or 2 μl of anti-GFP (A11122 Invitrogen) to solubilized chromatin and incubated overnight at 4°C. 25 μl of Protein G Dynabeads (Thermofisher) were added for 3H at 4°C. Beads were then washed twice with ice-cold Lysis Buffer Low Sodium, twice with ice-cold Lysis Buffer High Sodium (50 mM HEPES–KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-Deoxycholate), twice with ice-cold sodium deoxycolate buffer (10 mM Tris–HCl pH 8, 250mM LiCl, 0.5% NP-40, 0.5% Na-Deoxycholate, 1mM EDTA) and once with ice-cold TE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA). Beads were then eluted in TES (TE with 1% of SDS) and incubated for 15 min at 65°C. Samples were incubated 12 h at 65°C and then treated with Proteinase K during 4 h at 37°C. DNA was purified using innuPREP PCRpure purification column (Analytikjena) and samples were analyzed by qPCR.
qPCR and statistical analysis for ChIP
Input samples were diluted to 1/40. 2 μl of template DNA was added to 5 μl of TB Green Premix Ex Taq II (Takara), and primers were added to a final concentration of 0.4 μM. Each sample was run in triplicate on the same 96-well PCR plate in a C1000 Touch Thermal Cycler (CFX96 Biorad). PCR program was as followed: 95°C for 30 s followed by 55 cycles at 95°C for 10 s, 55 or 56°C (for 2R-48 bp or control 100 kb, respectively) for 10 s, 72°C for 10 s and a final extension step at 72°C for 5 min. Primers used for qPCR are listed in Supplementary Table S2. Corrected % of IP was calculated as followed: Corrected % IP = % IP 2R-48 bp – % IP 100 kb where 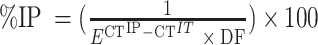 with E the efficiency of qPCR,
with E the efficiency of qPCR, 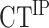 and
and 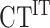 the mean of triplicate for IP and Input (IT) respectively, DF the dilution factor. Statistical comparisons were performed using a Mann–Whitney test when variance is significantly different between the two analyzed data sets or unpaired two-tail t-test if not. * P-value < 0.05, ** P-value < 0.01, *** P-value < 0.005.
the mean of triplicate for IP and Input (IT) respectively, DF the dilution factor. Statistical comparisons were performed using a Mann–Whitney test when variance is significantly different between the two analyzed data sets or unpaired two-tail t-test if not. * P-value < 0.05, ** P-value < 0.01, *** P-value < 0.005.
Western blot
Proteins were extracted with trichloracetic acid (TCA) from indicated strains, as previously described in Coulon et al. (41). Protein samples are loaded on 6% acrylamide gel and monitored using anti-flag (M2, mouse monoclonal antibody). Ponceau-S staining of the membrane was used as a loading control.
RESULTS
Telomerase elongates an eroded proto-telomere in G0
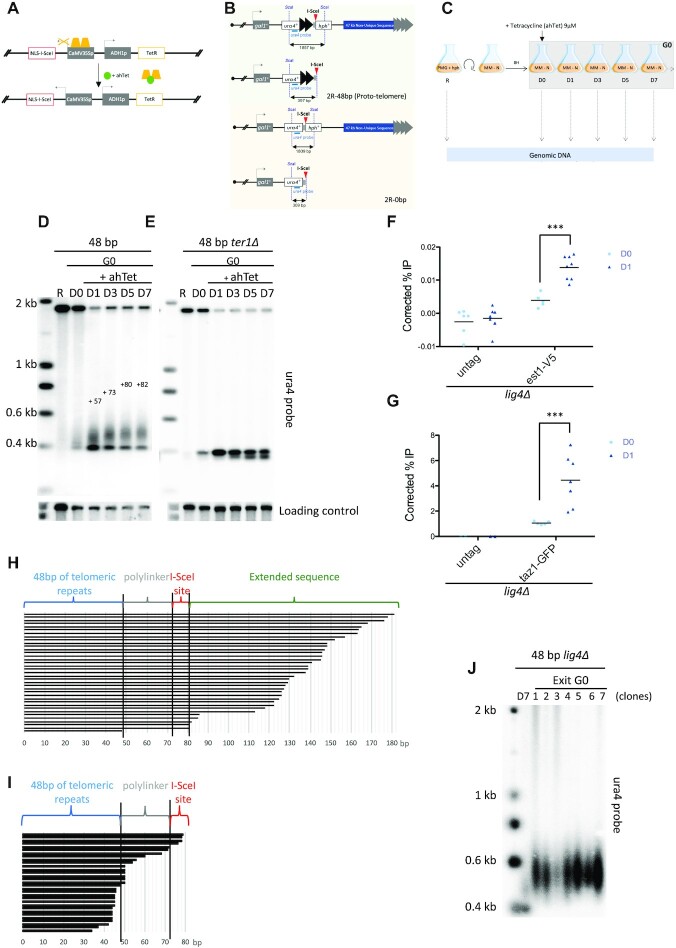
To investigate how eroded telomeres are processed in post-mitotic cells, we took advantage of the conditional telomere-repeat capped end system developed in the Runge laboratory (Figure 1A and B) (40). This artificial end, named proto-telomere, mimics in many aspects a single short telomere in cycling cells (42). Briefly, a single I-SceI cut site was integrated ∼47 kb away from the right telomere of chromosome II (C2-R) flanked by selectable markers (ura4+ and hph+). Upon induction of the endonuclease I-SceI with tetracycline (ahTet), a single I-SceI cut site is generated leading to the formation of the proto-telomere bearing either 48 bp of telomeric repeats or no repeats as a control (2R-48 bp and 2R-0 bp, respectively), marked with ura4+. Importantly, loss of the 47 kb of non-unique subtelomeric sequences in C2-R (containing an hph+ cassette) is dispensable for cell viability (40). In vegetative cells, the 2R-48bp is rapidly elongated by telomerase and reaches a final length of 250 bp after approximately 8 population doublings (PD), while the 2R-0 bp is degraded (40). To determine the fate of the proto-telomere in quiescent cells, we introduced this system in prototrophic fission yeast strains and induced I-SceI by adding ahTet once cells are arrested in G0 after nitrogen starvation. We collected cells before induction of I-SceI at day 0 (D0) and 1, 3, 5 and 7 days after induction (D1 to D7) (Figure 1C). For each experiment, we checked that the cell population was completely arrested in G0 by microscopic observation and FACS analysis (as exemplified by Supplementary Figure S1D). Genomic DNA was extracted for each time point, digested with ScaI, subjected to Southern blot analysis, and the 2R proto-telomere was revealed with an ura4 probe (Figure 1D). The uncut 1.9 kb ura4+::2R-48 bp::hph+ band was visible prior to induction of I-SceI and was replaced by the cut 0.4 kb ura4+::2R-48 bp band after induction. At D1, the elongation of the 2R-48 bp was already visible and it continued until D7. We reproducibly observed an elongation of approximately 80 bp (±5 bp) of the telomeric seed. Notably, the cut and uncut bands remained detectable during the time in quiescence (see below, Figure 2). To confirm that elongation was the result of telomerase action, we induced the 2R-48 bp in ter1Δ strain. No elongation was detected in the ter1Δ strain but a band just below the cut fragment appeared with time in G0 (Figure 1E). Importantly, we did not observe telomere elongation with the 2R-0 bp (Supplementary Figure S1A). Chromatin Immunoprecipitation (ChIP) of telomerase subunit Est1 showed significant enrichment at the 2R-48 bp after I-SceI induction (D1) (Figure 1F and Supplementary Figure S1B). Of note, ChIP experiments were performed in lig4Δ cells to avoid ligation of the cut product (see next paragraph). In addition, ChIP of Taz1-GFP confirmed the presence of Taz1 at the 2R-48 bp before the I-SceI cut and showed that Taz1 was enriched after the generation of the proto-telomere (Figure 1G). Sequencing of the elongated 2R-48bp fragments extracted from agarose gel revealed that an average of 63 bp of telomeric repeats were added to the proto-telomere (Figure 1H and Supplementary Figure S1C), slightly lower than that estimated by Southern blot. This could be explained by a bias during the telomere cloning step performed for telomere sequencing. Sequencing of the shortened 2R-48 bp fragments extracted from agarose gel in the ter1Δ strain revealed that the degradation of the I-SceI and polylinker sequence was observed in most cases, but importantly telomeric repeats were maintained (Figure 1I).
Figure 1.
The 2R-48bp is extended by telomerase in quiescence. (A) The DNA sequence-specific endonuclease I-SceI is expressed under the control of a tetracycline-inducible promoter (TetR-controlled CaMV35S promoter) in a cassette that also expresses TetR. The addition of tetracycline (ahTet) induces I-SceI expression, which then cuts at site introduced into the genome. (B) The proto-telomere cassette is integrated downstream of the gal1+ gene, ∼47 kb from the right telomere of chromosome II (C2-R). The 2R-48 bp proto-telomere contains the ura4+ gene followed by 48 bp of telomere repeats (black triangles), a polylinker sequence (grey square), the I-SceI site (red triangle) and the hygromycin resistance marker (hph+), while the 0bp control lacks the telomere repeats. The S. pombe native telomere repeat tracts are indicated by gray triangles. Relative position of the restriction sites and probe used for Southern Blot analysis are indicated. (C) Schematic workflow of a typical quiescence experiment. Cells were grown in minimal medium (PMG + hygromycin) then shifted in nitrogen-depleted medium (MM-N) and maintained in quiescence for seven days. ahTet was added 8 h after nitrogen starvation to induce the proto-telomere. Samples were collected at different time points as indicated to perform genomic DNA extraction. (D, E) Genomic DNA from quiescent WT and ter1Δ cells was digested with ScaI and Southern blotted. Membrane was hybridized with ura4 probe to reveal the cut and uncut proto-telomere (2R-48 bp). D1, D3, D5 and D7 correspond to number of days for which cells were in G0 after ahTet addition before collecting and R indicates replicative sample collected before nitrogen-starvation. The membrane was also hybridized with abo1 probe used as a loading control. In D, the size (bp) of extended 2R-48bp is indicated. (F, G) Scatter plot representation of chromatin immunoprecipitation (ChIP) of Est1-V5 and Taz1-GFP in lig4Δ cells containing the 2R-48bp proto-telomere cassette. Quiescent cells were collected before I-SceI induction (D0) and 12H hours after induction with ahTet (D1). The immunoprecipitated DNA was analyzed by quantitative PCR with primers located nearby the cut (2R-48bp) and at 100 kb distance from the I-SceI site (control). The corrected % IP is the percentage of immunoprecipitated DNA of the target minus the control locus. Each dot corresponds to an individual experiment. Statistical comparisons were performed using an unpaired two-tailed t-test and a Mann–Whitney two-tailed test for Est1-V5 and Taz1-GFP ChIP, respectively (*** P-value < 0.005). (H, I) Schematic alignment of sequences obtained after 7 days in G0 (D7). For H the extended 2R-48 bp shown in panel D was gel extracted, cloned and sequenced. For I the shortened 2R-48 bp (the lower band of panel E) was gel extracted, cloned and sequenced. (J) After 7 days of nitrogen starvation in the presence of ahTet, G0 cells shown in panel D (2R-48 bp) were micromanipulated on rich YES-plates to allow exit from quiescence. Genomic DNA was extracted from seven individual clones, digested with ScaI and Southern blotted. Sample D7 of quiescence is loaded to compare the proto-telomere elongation to other samples. Membrane was hybridized with ura4 probe. Further extension of the 2R-48 bp was observed after cell cycle re-entry.
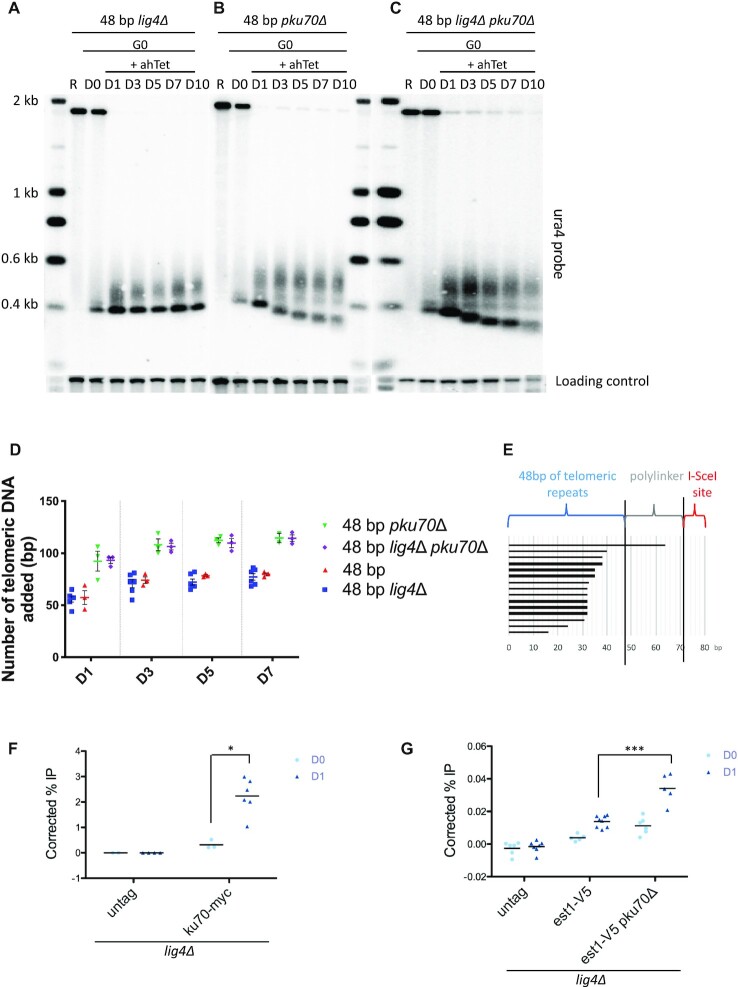
Figure 2.
The NHEJ pathway and telomerase both process the proto-telomere in quiescence. (A–C) Genomic DNA from quiescent lig4Δ, pku70Δ and lig4Δ pku70Δ cells was digested with ScaI and Southern blotted. Membrane-bound DNA was hybridized with ura4 and abo1 probes. R (replicating cells), D0 (G0 before ahTet addition), D1, D3, D5, D7 and D10 (days in G0 after addition of ahTet) samples were collected. (D) Quantification of proto-telomere extension from panel A, B, C, and Figure 1D. Individual values are plotted from at least three different experiments, the colored and the black lines indicating the mean and the SEM, respectively. (E) Schematic alignment of sequences obtained from the shortest ura4+::2R-48 bp in pku70Δ cells after 7 days in G0 (D7). The lower band in panel B was gel extracted, cloned and sequenced. (F, G) Chromatin immunoprecipitation (ChIP) of Ku70-myc and Est1-V5 in lig4Δ cells containing the 2R-48bp proto-telomere cassette. Quiescent cells were collected before I-SceI induction (D0) and 12H hours after induction with ahTet (D1). The immunoprecipitated DNA was analyzed by quantitative PCR with primers located nearby the cut (2R-48 bp) and 100 kb away from the I-SceI site (control). The corrected % IP was plotted for each sample, and the values obtained in individual experiments are shown. Statistical comparisons were performed using an unpaired Mann–Whitney two-tailed test for Ku70-myc and Est1-V5ChIP (* P-value < 0.05 and *** P-value < 0.005).
From these results, we concluded that telomerase is able to elongate the short proto-telomere generated by the I-SceI cut in quiescence, despite the absence of cell cycle-dependent control and the passage of the replication fork. Importantly, in quiescence the proto-telomere reached a final length of ∼130 bp after extension by the telomerase (48 bp from the seed plus the 80 bp of extension), a size significantly shorter than that produced by 2R-48bp extension in vegetative cells (250 bp) (40). Figure 1J shows that when cells undergo exit from G0 and cell cycle re-entry they further extend the 2R-48 bp up to the size of 180–250 bp, revealing a steady state size difference between G0 and cycling cells. This size difference might reveal the existence of a negative feedback control of telomerase action in G0 or might indicate that telomerase is less active in quiescent than in vegetative cells.
Notably, in most of our experiments we observed that a small fraction of cut fragment appeared before the induction of I-SceI by ahTet (Figure 1D). We confirmed that early cleavage occurred when cells enter into quiescence by monitoring ura4+-2R-48 bp directly after nitrogen starvation (Supplementary Figure S1D). This observed premature cleavage was likely caused by the negative control of adh1 promoter that occurred upon nitrogen starvation, leading to decreased level of tetracycline repressor (34). Indeed, in the absence of I-SceI no cleavage was observed (Supplementary Figure S1E). For this reason, we wanted to induce I-SceI expression as soon as cells are arrested, 6–8 h after starvation as previously described (43). FACS analysis confirmed that cells fully entered into quiescence after 8 hours (Supplementary Figure S1D), thus we decided to induce the cut after 8 hours of starvation in the following experiments.
Both NHEJ and telomerase act upon the 48 bp proto-telomere
In fission yeast arrested in G0, NHEJ is a prominent pathway to repair DSB (44–46). We wondered whether a competition exists between the elongation of the proto-telomere by telomerase and the repair of the I-SceI break by NHEJ. We thus analyzed the fate of the proto-telomere in cells lacking Lig4 and Ku70, two main players of the NHEJ pathway. In the absence of Lig4, the 2R-48bp was elongated and the ura4+::2R-48 bp was stabilized over the G0 period (Figure 2A). The uncut band completely disappeared at D1 in lig4Δ cells, in contrast to what observed in lig4+ cells (Figure 1D), indicating that NHEJ was efficiently acting upon the proto-telomere in quiescence. I-SceI digestion of the ura4+::2R-48 bp::hph+ PCR-amplified fragments revealed that a fraction of this fragment become resistant to I-SceI cleavage (Supplementary Figure S2A), indicating that error-prone NHEJ occurs in lig4+ cells, and explaining the persistence of the uncut fragment observed in Figure 1D (47). Despite the fact that I-SceI cleaved more efficiently the ura4+::2R-48bp::hph+ in lig4Δ cells, the extent of elongation by telomerase remained similar to that in lig4+ cells (Figure 2A and D). We next investigated the impact of Ku complex on the elongation of the 2R-48 bp (Figure 2B). In the absence of Ku70, the uncut fragment progressively shortens, likely due to exonuclease activity, indicating that NHEJ was inefficient similarly to that in lig4Δ cells. Importantly, the length of the extended 2R-48bp increased compared to pku70+ cells (Figure 2D) with the concomitant gradual degradation of the cut fragment (Figure 2B). In line with this, we also observed degradation of the 2R-0 bp (Supplementary Figure S2B). Taken together, these results suggest that the Ku heterodimer promotes NHEJ, protects the extremity of the proto-telomere, and prevents telomerase action. Sequencing of the shortest ura4+::2R-48 bp fragment extracted from agarose gel at D7 in pku70Δ cells confirmed the shortening of the telomeric seed (Figure 2E).
To confirm the presence of Ku at the 2R-48 bp, we performed ChIP of myc-tagged Ku70 protein at the 2R-48 bp in lig4Δ cells. We observed a significant enrichment of Ku70 after I-SceI induction (Figure 2F). Consistent with this result, telomerase was more enriched at the proto-telomere in the absence of Ku70 compared to WT strain (Figure 2G and Supplementary Figure S1B). These observations indicate that the Ku complex protects the proto-telomere from degradation and at the same time hampers telomere extension by telomerase. Deleting lig4+ in pku70Δ cells did not further modify the I-SceI cleavage pattern or the 2R-48 bp length (Figure 2C and D).
Overall, our results indicate that Ku is required to promote NHEJ that is the prominent pathway operating at the 2R-48 bp. Telomerase is also able to elongate the proto-telomere in a fraction of cells, however the absence of Ku does not increase the number of cells able to elongate the proto-telomere. This result indicates that Ku interferes with the processivity of telomerase engaged with the proto-telomere, but does not affect the initiation step. Based on these observations, we decided to monitor the fate of the proto-telomere in the absence of Lig4 in the following experiments to avoid the interference of the NHEJ.
Rad3ATR controls telomerase activity through Ccq1 phosphorylation
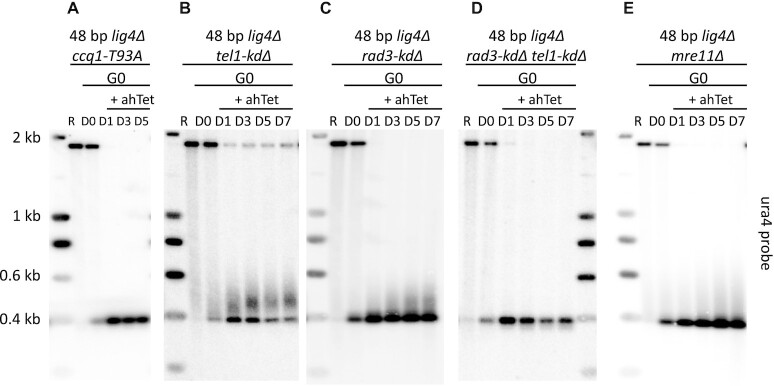
In cycling cells, telomerase recruitment to telomeres is restricted to late S/G2 phase and telomere elongation is intimately linked to replication. Rad3ATR and Tel1ATM phosphorylate Ccq1 on threonine 93 (T93), Rad3ATR being the prominent kinase involved in telomerase activation. In the absence of replication, the question of telomerase activity regulation arises. We found that the 2R-48bp was not elongated in ccq1-T93A cells (Figure 3A), indicating that Ccq1 phosphorylation is a crucial step for telomerase activation in quiescent cells. Next, we monitored elongation of the proto-telomere in mutants of rad3+ and tel1+ carrying a deletion of the kinase domain (rad3-kdΔ, tel1-kdΔ and rad3-kdΔ tel1-kdΔ double mutants). While the elongation of the 2R-48bp was not affected in tel1-kdΔ (Figure 3B), extension of the proto-telomere was reduced in rad3-kdΔ cells (Figure 3C), and completely abolished in the rad3-kdΔ tel1-kdΔ double mutant as in ccq1-T93A cells (Figure 3D). We concluded that both Rad3ATR and Tel1ATM are important to control the elongation of the proto-telomere in G0, Rad3ATR being the prominent kinase such as in cycling cells (48,49).
Figure 3.
Rad3ATR controls telomerase activity through Ccq1 phosphorylation. (A–E) Genomic DNA from quiescent ccq1-T93A lig4Δ, tel1-kdΔ lig4Δ, rad3-kdΔ lig4Δ , rad3-kdΔ tel1-kdΔ lig4Δ and mre11Δ lig4Δ cells was digested with ScaI and Southern blotted. Membrane was hybridized with ura4 probe. R (replicating cells), D0 (G0 before ahTet addition), D1, D3, D5 and D7 (days in G0 after addition of Tc) samples were collected.
Current models propose that ATR-ATRIP is recruited to DNA damage sites by interacting with replication protein A (RPA)-covered single-stranded DNA (ssDNA) (50,51). By analogy, Rad3ATR-Rad26ATRIP is thought to be recruited by RPA at telomeres to phosphorylate Ccq1 during telomere replication in fission yeast (48). Because the proto-telomere extension mainly relies on Rad3 in G0 cells, it suggests that RPA-coated ssDNA, generated by resection of the 2R-48 bp could be a key step for the proto-telomere extension. We were unable to demonstrate by ChIP the enrichment of Rpa1 at the 2R-48 bp after induction of the cut. However, we showed that Mre11, a member of the MRN complex that is important to initiate resection, likely by clipping Ku from double-stranded DNA (52,53), was indeed required for the proto-telomere extension in quiescent cells (Figure 3E). This suggests that Mre11-dependent resection of the 2R-48bp and the eviction of Ku are important to promote proto-telomere extension.
Taz1 and Rap1 are required for proto-telomere elongation in quiescent cells
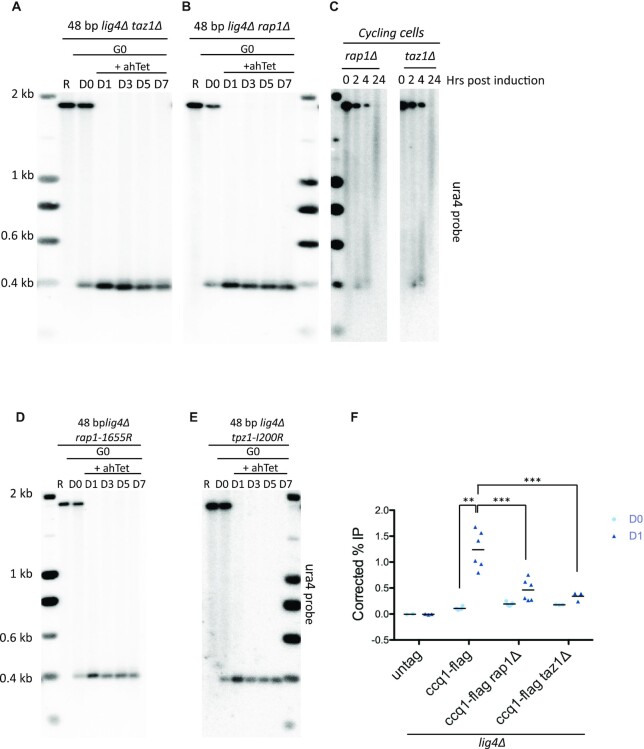
In cycling cells, loss of Taz1 or Rap1 results in highly elongated telomeres, indicating that these factors inhibit telomere elongation by controlling telomerase activity (37,38). Hence, we investigated the involvement of these telomeric proteins in the 2R-48 bp elongation. Strikingly, the absence of Taz1 or Rap1 strongly impaired the extension of the proto-telomere in quiescent cells since only a limited extension of the proto-telomere was detected in a small fraction of cells (Figure 4A and B). This is in contrast with the situation in vegetative cells where Taz1 and Rap1 act as negative regulators of telomerase. We demonstrated that by showing that the proto-telomere was indeed overextended when the cut was induced in taz1Δ and rap1Δ cycling cells (Figure 4C). Therefore, the role that both Taz1 and Rap1 play during extension of the proto-telomere is different in vegetative and quiescent cells.
Figure 4.
Taz1 and Rap1 positively regulate telomerase activity in quiescent cells. (A-B,D-E) Genomic DNA from quiescent taz1Δ lig4Δ , rap1Δ lig4Δ, rap1-I655R lig4Δ and tpz1-I200R lig4Δ cells was digested with ScaI and Southern blotted. Membrane was hybridized with ura4 probe. R (replicating cells), D0 (G0 before ahTet addition), D1, D3, D5 and D7 (days in G0 after addition of ahTet) samples were collected. (C) Analysis of the 2R-48bp extension in taz1Δ and rap1Δ vegetative cells after ahTet addition. Genomic DNA was extracted, digested with ScaI and Southern blotted. Membrane was hybridized with ura4 probe. (F) Ccq1-Flag ChIP in WT, rap1Δ and taz1Δ lig4Δ cells containing the 2R-48 bp proto-telomere cassette. Quiescent cells were collected before (D0) and 12H hours after (D1) I-SceI induction with ahTet. The immunoprecipitated DNA was analyzed by quantitative PCR with primers located nearby the cut (2R-48 bp) and 100 kb away from the I-SceI site (control). The corrected % IP was plotted with the values obtained in individual experiments shown for each sample. Statistical comparisons were performed using an unpaired Mann–Whitney two-tailed test to compare D0 and D1 of Ccq1-flag in WT and an unpaired two-tailed t-test for the others ** P-value < 0.01 and *** P-value < 0.005).
Because the proto-telomere lacks a canonical telomeric overhang, we speculated that the function of Taz1 is to promote the shelterin assembly at the cut site. To test this hypothesis, we analyzed the fate of the proto-telomere in rap1-I655R and tpz1-I200R mutants which are deficient for the interaction between Taz1 and Pot1, respectively (54,55). In both mutants, the extension of the 2R-48 bp is impaired (Figure 4D and E), showing that the shelterin bridge is crucial for elongation of the proto-telomere by telomerase.
To strengthen the role played by both Taz1 and Rap1 in promoting telomerase action in G0, we further monitored the recruitment of Ccq1 protein at the 2R-48bp (Figure 4F). After the induction of the I-SceI cut, a strong recruitment of Ccq1 protein was observed, which was significantly reduced in the absence of Taz1 and Rap1. These data confirm the role of Taz1-Rap1 in G0 for the proto-telomere elongation, and the need of the presence of Taz1-Rap1 to recruit Ccq1. Based on our data, we speculate that the role of Taz1 and Rap1 is to recruit the other telomeric elements to allow telomerase recruitment at the 2R-48 bp. Remarkably, Taz1 and Rap1 play different roles in the proto-telomere elongation in cycling and quiescent cells.
Stn1 and Rif1 negatively regulate proto-telomere extension
We next analyzed the role of the Stn1-Ten1 complex that inhibits telomerase action by promoting the synthesis of the complementary strand and whose recruitment to telomere occurs through its interaction with the SUMO-modified Tpz1 (56–59). The stn1-226 allele carries point mutations in the SIM domain of Stn1 that provokes the telomerase-dependent elongation of telomeres (57). In line with these results, we observed that in stn1-226 cells, the 2R-48bp was overelongated compared to stn1+ cells (Figure 5A). Our data indicate that inhibition of telomere elongation is similarly controlled by Stn1-Ten1 complex in quiescence.
Figure 5.
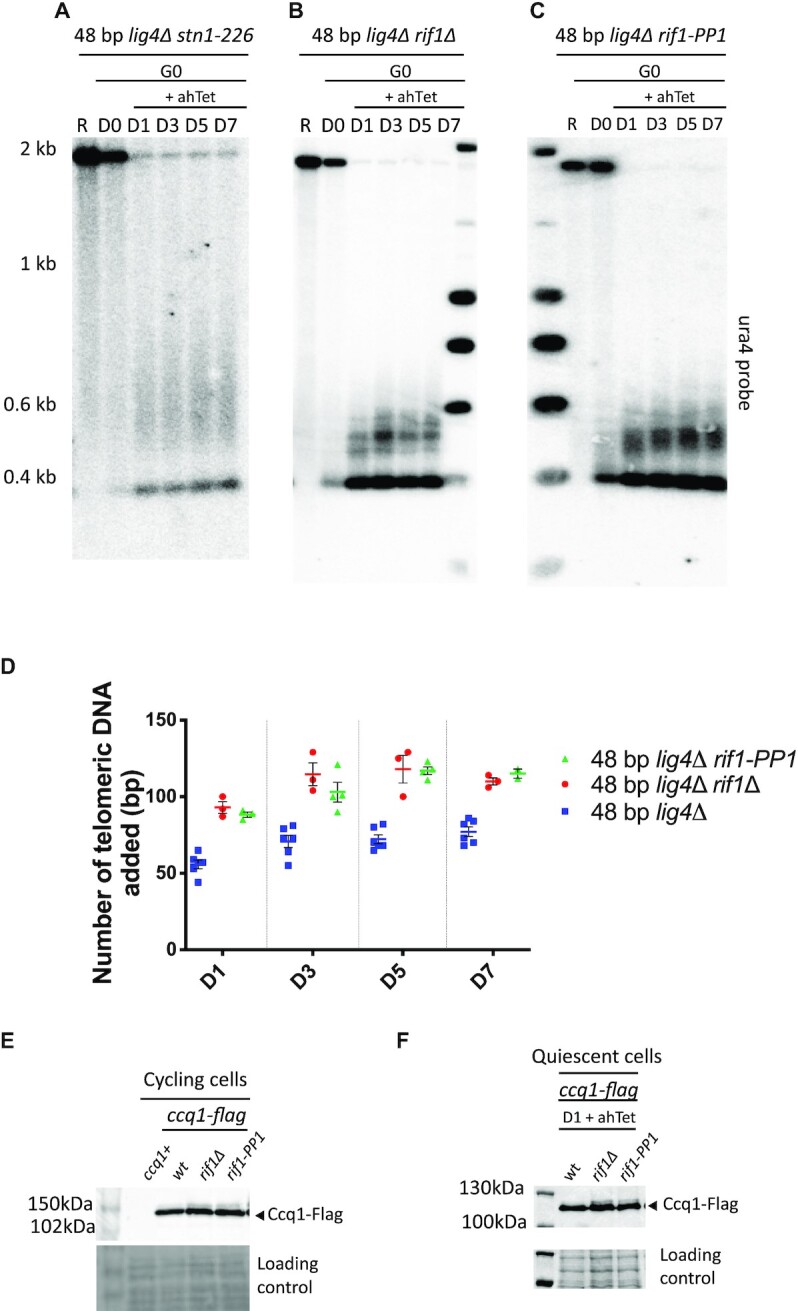
Stn1 and Rif1 negatively regulate telomerase activity in quiescent cells. (A–C) Genomic DNA from quiescent snt1-226 lig4Δ, rif1Δ lig4Δ, and rif1-PP1 lig4Δ cells was digested with ScaI and Southern blotted. Membrane was hybridized with ura4 probe. R (replicating cells), D0 (G0 before ahTet addition), D1, D3, D5 and D7 (days in G0 after addition of ahTet) samples were collected. (D) Quantification of proto-telomere extension shown in panel B, C and Figure 2A. Individual values are plotted from at least 3 different experiments, the colored and the black lines indicating the mean and the SEM, respectively. (E) The phosphorylation of Ccq1 from indicated strains carrying the 2R-48bp was assessed by Western blot. TCA extraction was performed from vegetative cells before nitrogen starvation and immunoblotted with anti-flag antibody. The slow-migrating band corresponds to phosphorylated Ccq1. Ponceau red staining was used as loading control. (F) The phosphorylation of Ccq1 from indicated strains carrying the 2R-48 bp was assessed by western blot. TCA extraction was performed from cell samples maintained one day in quiescence with addition of ahTet to induce the I-SceI cut.
Rif1 is also a negative regulator of telomerase activity (38). Thus, we further investigated the involvement of Rif1 in the control of telomere elongation in G0. Rif1 controls the timing of telomere replication by repressing the firing of late origins via its associated PP1 phosphatases (60–62). As a consequence, early replication of telomeres in cycling cells has been proposed to affect the length of telomeres (23). We found that the 2R-48 bp was also overelongated in rif1Δ quiescent cells (Figure 5B and D) in conditions in which replication origins are dormant. Moreover, in rif1-PP1 cells expressing a Rif1 mutant form unable to bind PP1 phosphatases (63), the 2R-48 bp was extended as in rif1Δ cells (Figure 5C and D). These results suggest that Rif1 regulates telomere-length in quiescence through its associated PP1 phosphatases. We thus hypothesized that Rif1-PP1 may control telomerase activity by dephosphorylating Ccq1. Hence, we monitored in vegetative and in quiescent cells the phosphorylation of Ccq1 in rif1Δ and rif1-PP1 mutants. As previously reported (39), we confirmed that phosphorylation of Ccq1 was stabilized in rif1Δ cells and also in the rif1-PP1 mutant (Figure 5E). Strikingly, we were also able to detect Ccq1 phosphorylation stabilization after induction of I-SceI in rif1Δ and rif1-PP1 quiescent cells (Figure 5F).
Ccq1 is phosphorylated at multiple sites including the T93 residue known to control telomerase recruitment (30,31). We could not establish whether Rif1/PP1 directly controls the dephosphorylation of Ccq1 T93 amino acid, however our results indicate that Rif1/PP1 is responsible for Ccq1 dephosphorylation in both vegetative and quiescent cells in which it controls the proto-telomere extension. Our results also point to the molecular mechanism by which Rif1 negatively regulates telomerase activity in fission yeast in general.
DISCUSSION
In this study, we investigated in quiescent fission yeast cells the fate of a telomere-repeat capped end. This proto-telomere mimics a native single short telomere in many aspects in cycling cells (40,42). Like in cycling cells, the presence of the telomeric seed (48 bp) near the break promotes de novo telomere addition despite the fact that the I-SceI 3′-overhang does not exhibit telomeric sequences. This is reminiscent to HO-cleavage adjacent to a telomeric seed sequence which has been extensively exploited to study telomere elongation in budding yeast (64). We further report that telomerase can extend this short proto-telomere despite that telomere elongation was thought to depend on replication fork passage in most organisms studied (18,22,23). Thus, the extension of the proto-telomere by telomerase can occur outside of S-phase in the absence of replication fork passage in quiescent cells, likely with a different kinetic due to the low resection activity in arrested cells (65). Our findings were reminiscent of previous observations in fission yeast showing that telomere elongation was observed in G1 in taz1Δ cells. However, in those cells lacking the main shelterin component Taz1, telomerase access to telomeres was no longer cell-cycle restricted and cells accumulated structures that can serve as substrates for telomerase (66). Taken together, the extension of a proto-telomere in arrested cells indicates that telomerase could also elongate native short telomeres independently of the S-phase in quiescence.
In haploid cells, NHEJ becomes the prominent pathway of DSB repair in G1 phase. In S. pombe, Taz1 and Rap1 protect telomeres from Ku-dependent fusion in nitrogen-starved cells (46,67). In line with these studies, we report here that the Ku complex is recruited at the proto-telomere, protects it from degradation, and favors NHEJ (Figure 6A). As a consequence, removal of the Ku complex leads to an increased processivity of the 2R-48 bp extension indicating that Ku prevents telomerase action in non-dividing cells. We thus speculate that during elongation of the proto-telomere, telomerase can fall off and Ku binding can prevent further extension by telomerase (Figure 6C) (50,63). We further observed that the proto-telomere extension requires Mre11. The MRN complex is known to evict Ku from double-stranded DNA extremities and to initiate resection (53,68). These results suggest that the MRN complex triggers telomere elongation by promoting resection at the I-SceI break thereby creating a longer overhang (Figure 6A). We have previously established in cycling cells in which NHEJ is inhibited in favor of the HR pathway that Ku and telomerase can compete for the binding of telomeric DNA structures occurring during replication of DNA ends (69). A different regulation seems to occur in G0-arrested cells, since the fate of the 2R-48 bp, rejoining or extension, would rather depend on Ku activity. Whether or not Ku promotes NHEJ in quiescence has not been addressed here, but by analogy to DSB repair mechanism in cycling cells (47), we can speculate that Ku may promote NHEJ in arrested cells.
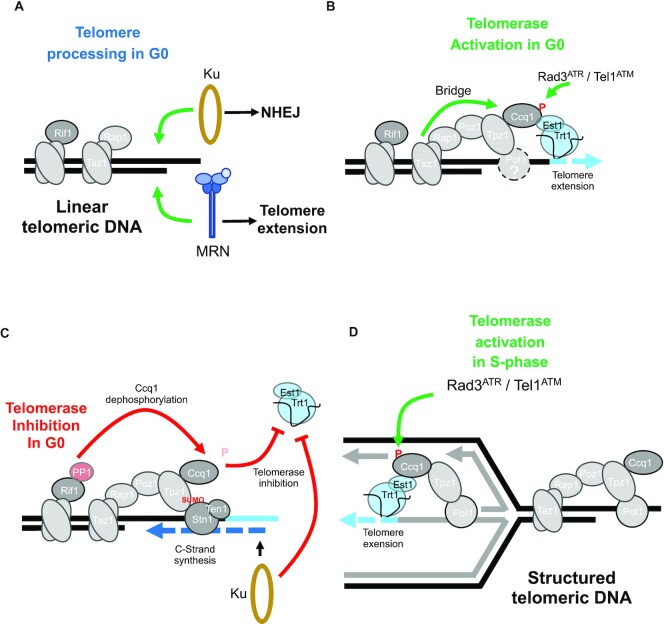
Figure 6.
Control of telomerase activity in vegetative and quiescent cells. (A) Telomere processing in quiescent cells. A short telomere is either recognized by Ku complex that will promote NHEJ or extended by telomerase. NHEJ is the most proficient pathway in G0 cells, however a telomeric seed is able to promote telomerase recruitment. (B) Telomerase activation in quiescent cells. Taz1 and Rap1 are both required to assemble shelterin via their bridging function to promote telomerase recruitment that requires the Rad3ATR/Tel1ATM-dependent phosphorylation of Ccq1. (C) Telomerase inhibition in quiescent cells. Inhibition of telomerase relies on the dephosphorylation of Ccq1 through Rif1-associated PP1 phosphatases and on the concomitant sumolylation of Tpz1 that promotes Stn1-Ten1 recruitment and synthesis of the complementary strand. Ku binding to the extended telomere is also thought to inhibit telomerase action. (D) Telomerase activation in cycling cells. Replication stress at short telomeres generates robust substrates, such as reversed forks, for telomerase. RPA-coated ssDNA promotes the Rad3ATR/Tel1ATM-dependent phosphorylation of Ccq1 which then recruits telomerase independently of Taz1-Rap1 shelterin assembly.
In this study, we show that Rad3ATR and Tel1ATM are redundantly required for the short telomere elongation in quiescence, likely by phosphorylating Ccq1 at position T93 (Figure 6B). Strikingly, in contrast to vegetative cells in which Taz1 and Rap1 negatively regulate telomerase-dependent elongation of the proto-telomere, they were both required for its extension in quiescence. Hampering the interaction between Taz1 and Rap1 or between Tpz1 and Pot1 also alter the extension of the proto-telomere. Taken together, this unveils crucial functions of the two shelterin elements Taz1 and Rap1 in promoting telomerase recruitment and telomerase activity in non-dividing cells. These results also suggest that the binding of Taz1 and Rap1 to the 48 bp telomeric seed might be essential to position Ccq1 and telomerase, likely through the bridging function of telomeric factors (Figure 6B). The question arises about the mechanism by which telomerase is recruited to the proto-telomere and how it engages with the 3' single-strand end. ChIP experiments in cycling cells reveal that Taz1 is already present at the proto-telomere before the cutting and that Pot1 is recruited later, after induction of the cut (42). Thus, it is likely that Pot1 is not initially present at the ISce-I break in G0 cells and that telomerase is positioned at the tip of the proto-telomere only through its interaction with the shelterin-bridged Ccq1 protein. However, Tpz1-Pot1 interaction is important for proto-telomere extension revealing the complexity of the function of Pot1 in controlling telomerase activity (26,70). When the proto-telomere is extended, it is likely that the fully assembled shelterin adopts then a closed state that could inhibit telomerase (71).
In vegetative cells, telomerase access and recruitment to telomeres are restricted by shelterin proteins to late-S/G2 phase (72,73). It has been also reported that stalled replication forks are likely to generate robust substrates for telomerase (66,69). Thus, in cycling cells two modes of action of telomerase possibly co-exist. The first may rely on the recruitment of telomerase to DNA structures generated during telomere replication and would be independent of Taz1-Rap1 bridge formation (Figure 6D). In line with this, in the absence of Taz1 or Rap1, replication stress increases and reversed forks accumulate providing DNA ends and ssDNA promoting telomerase recruitment at telomeres (37,38). The second mechanism of telomerase action would occur at telomeric linear extremities once replication is completed and would require Taz1 and Rap1 proteins to recruit telomerase to its site of action. As mentioned above, in quiescent cells, only the latter mechanism exists (Figure 6B). This could also explain the reduced lengthening of the proto-telomere in between vegetative and quiescent cells. Whatever the case, both modes of telomere extension depend on the Rad3/Tel1 phosphorylation of Ccq1. Whether elongation of the 2R-48 bp in quiescent cells depends on resection and on recruitment of Rad3ATR through RPA-coated ssDNA filament accumulation remains to be determined.
Rif1 controls the timing of telomere replication by repressing the firing of late origins through its binding to PP1 phosphatases in both budding and fission yeasts (60–62,74,75). It has been proposed that the effect of Rif1 on telomere length might be mediated through its effect on nearby replication origins. Earlier replication could potentially increase the window to recruit telomerase in S. cerevisiae (23). However, the function of Rif1 in origin firing was recently proposed to be separable from its role in telomere length regulation and independent of its PP1 phosphatase activity (76). In contrast, evidence showed that Rif1 may rather act through PP1 phosphatases to limit telomere elongation (62). In line with this latter study, we report here that Rif1 negatively regulates telomerase through its PP1 phosphatase activity by antagonizing Ccq1 phosphorylation in both vegetative and quiescent fission yeast cells (Figure 6C). This indicates that in S. pombe negative regulation of telomere length by Rif1 is independent of replication timing although we can’t rule out completely this possibility since rif1-PP1 mutant displays slightly shorter telomere than rif1Δ cycling cells (57). Nevertheless, the impact of replication timing seems to have a minor effect and the difference between rif1-PP1 and rif1Δ in telomere length can be attributed to a residual PP1 binding capacity of Rif1-PP1 protein (63). Moreover, we identify Ccq1 as a potential target of Rif1-associated PP1 phosphatases, linking Rif1 function to telomerase inhibition. Our results therefore unveil unforeseen insights in the control of telomerase activity in fission yeast. Indeed, dephosphorylation of Ccq1 by Rif1-associated PP1 phosphatases and concomitant Tpz1 sumolylation by Pli1 SUMO ligase as well as Stn1 dephosphorylation by Ssu72 phosphatase, all contribute to telomerase inhibition in cycling cells (56,58,59), and perhaps in quiescent cells as well.
We previously reported that telomerase can repair short telomeres in cycling cells using fission yeast as a model system (69). Here, we further uncovered that telomerase is able to elongate a single-short proto-telomere in quiescent cells in a way that shares similarities but also differences with the mode of action of telomerase in replicating cells. Although many questions still remain, our study paves the way for the understanding of the telomere maintenance mechanism in post-mitotic cells.
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the pombe community for strains, in particular Junko Kanoh, Toru Nakamura, Feng Qiao and Julie Cooper. We also thank Benoit Arcangioli, Bertrand Llorente, Miguel Ferreira and the members of V.G. team for helpful discussions, in particular Dimitri Churikov for reading this manuscript and Christelle Cayrou. We are very grateful to Samah Matmati and Laetitia Maestroni, former members of the team, for technical assistance.
Author contribution: M.V. performed all the experiments presented in this study. M.V., V.G. and S.C. designed experiments and analyzed the data. J.A. and K.R. provided the inducible short telomere system and expertise. M.V, S.C. and V.G. wrote the paper.
Contributor Information
Mélina Vaurs, CNRS, INSERM, Aix Marseille Univ, Institut Paoli-Calmettes, CRCM, Equipe labellisée par la Ligue Nationale contre le Cancer, Marseille, F-13009, France.
Julien Audry, Department of Inflammation and Immunity, Lerner Research Institute, Cleveland Clinic Foundation, Cleveland, OH, USA.
Kurt W Runge, Department of Inflammation and Immunity, Lerner Research Institute, Cleveland Clinic Foundation, Cleveland, OH, USA; Department of Genetics and Genome Sciences, Case Western Reserve University, Cleveland, OH, USA.
Vincent Géli, CNRS, INSERM, Aix Marseille Univ, Institut Paoli-Calmettes, CRCM, Equipe labellisée par la Ligue Nationale contre le Cancer, Marseille, F-13009, France.
Stéphane Coulon, CNRS, INSERM, Aix Marseille Univ, Institut Paoli-Calmettes, CRCM, Equipe labellisée par la Ligue Nationale contre le Cancer, Marseille, F-13009, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
J.A. and K.R. are supported by US National Science Foundation [1908875]; US National Institutes of Health [AG051601]; V.G. laboratory is supported by the ‘Ligue Nationale Contre le Cancer’ (LNCC) (Equipe labélisée); M.V. is supported by ‘Bourse du Ministère de l’Enseignement Supérieur, et de la Recherche (MESR)'; S.C. is supported by the Agence Nationale de la Recherche [ANR-16-CE12 TeloMito, ANR-20-CE12 TeloRPA]. Funding for open access charge: Agence Nationale de la Recherche (ANR).
Conflict of interest statement. None declared.
REFERENCES
- 1. Lange T.de Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018; 52:223–247. [DOI] [PubMed] [Google Scholar]
- 2. Higa M., Fujita M., Yoshida K.. DNA replication origins and fork progression at mammalian telomeres. Genes (Basel). 2017; 8:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maestroni L., Matmati S., Coulon S.. Solving the telomere replication problem. Genes. 2017; 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taddei A., Gasser S.M.. Structure and function in the budding yeast nucleus. Genetics. 2012; 192:107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lingner J., Cooper J., Cech T.. Telomerase and DNA end replication: no longer a lagging strand problem?. Science. 1995; 269:1533–1534. [DOI] [PubMed] [Google Scholar]
- 6. Artandi S.E., Cooper J.P.. Reverse transcribing the code for chromosome stability. Mol. Cell. 2009; 36:715–719. [DOI] [PubMed] [Google Scholar]
- 7. Lansdorp P. Telomere length regulation. Frontiers Oncol. 2022; 12:943622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shay J.W., Wright W.E.. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 2010; 584:3819–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greenberg R.A., Allsopp R.C., Chin L., Morin G.B., DePinho R.A.. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998; 16:1723–1730. [DOI] [PubMed] [Google Scholar]
- 10. Holt S.E., Wright W.E., Shay J.W.. Regulation of telomerase activity in immortal cell lines. Mol. Cell. Biol. 1996; 16:2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daniali L., Benetos A., Susser E., Kark J.D., Labat C., Kimura M., Desai K., Granick M., Aviv A.. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun. 2013; 4:1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mamdani F., Rollins B., Morgan L., Myers R.M., Barchas J.D., Schatzberg A.F., Watson S.J., Akil H., Potkin S.G., Bunney W.E.et al.. Variable telomere length across post-mortem human brain regions and specific reduction in the hippocampus of major depressive disorder. Transl Psychiatry. 2016; 5:e636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aeby E., Ahmed W., Redon S., Simanis V., Lingner J.. Peroxiredoxin 1 protects telomeres from oxidative damage and preserves telomeric DNA for extension by telomerase. Cell Rep. 2016; 17:3107–3114. [DOI] [PubMed] [Google Scholar]
- 14. Fouquerel E., Lormand J.D., Bose A., Lee H.-T., Kim G.S., Li J., Sobol R.W., Freudenthal B.D., Myong S., Opresko P.L.. Oxidative guanine base damage regulates human telomerase activity. Nat. Struct. Mol. Biol. 2016; 23:1092–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kosmadaki M.G., Gilchrest B.A.. The role of telomeres in skin aging/photoaging. Micron. 2004; 35:155–159. [DOI] [PubMed] [Google Scholar]
- 16. Fouquerel E., Barnes R.P., Uttam S., Watkins S.C., Bruchez M.P., Opresko P.L.. Targeted and persistent 8-oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol. Cell. 2019; 75:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan A., Boulé J.-B., Zakian V.A.. Two pathways recruit telomerase to saccharomyces cerevisiae telomeres. PLos Genet. 2008; 4:e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diede S.J., Gottschling D.E.. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999; 99:723–733. [DOI] [PubMed] [Google Scholar]
- 19. Taggart A.K.P. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002; 297:1023–1026. [DOI] [PubMed] [Google Scholar]
- 20. Verdun R.E., Crabbe L., Haggblom C., Karlseder J.. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 2005; 20:551–561. [DOI] [PubMed] [Google Scholar]
- 21. Zhao Y., Sfeir A.J., Zou Y., Buseman C.M., Chow T.T., Shay J.W., Wright W.E.. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009; 138:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dionne I., Wellinger R.J.. Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 1998; 26:5365–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greider C.W. Regulating telomere length from the inside out: the replication fork model. Genes Dev. 2016; 30:1483–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coller H.A. The essence of quiescence. Science. 2011; 334:1074–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bighetti-Trevisan R.L., Sousa L.O., Castilho R.M., Almeida L.O.. Cancer stem cells: powerful targets to improve current anticancer therapeutics. Stem Cells International. 2019; 2019:9618065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyoshi T., Kanoh J., Saito M., Ishikawa F.. Fission yeast pot1-tpp1 protects telomeres and regulates telomere length. Science. 2008; 320:1341–1344. [DOI] [PubMed] [Google Scholar]
- 27. Dehé P.-M., Cooper J.P.. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 2010; 584:3725–3733. [DOI] [PubMed] [Google Scholar]
- 28. Moser B.A., Nakamura T.M.. Protection and replication of telomeres in fission yeast. Biochem. Cell Biol. 2009; 87:747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomita K., Cooper J.P.. Fission yeast ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008; 22:3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moser B.A., Chang Y.-T., Kosti J., Nakamura T.M.. Tel1ATM and Rad3ATR kinases promote ccq1-est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 2011; 18:1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamazaki H., Tarumoto Y., Ishikawa F.. Tel1(ATM) and rad3(atr) phosphorylate the telomere protein ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012; 26:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yanagida M. Cellular quiescence: are controlling genes conserved?. Trends Cell Biol. 2009; 19:705–715. [DOI] [PubMed] [Google Scholar]
- 33. Hassine S.B., Arcangioli B.. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J. 2009; 28:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bähler J.. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012; 151:671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maestroni L., Audry J., Matmati S., Arcangioli B., Géli V., Coulon S.. Eroded telomeres are rearranged in quiescent fission yeast cells through duplications of subtelomeric sequences. Nat. Commun. 2017; 8:1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maestroni L., Reyes C., Vaurs M., Gachet Y., Tournier S., Géli V., Coulon S.. Nuclear envelope attachment of telomeres limits TERRA and telomeric rearrangements in quiescent fission yeast cells. Nucleic Acids Res. 2020; 48:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cooper J.P., Nimmo E.R., Allshire R.C., Cech T.R.. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997; 385:744–747. [DOI] [PubMed] [Google Scholar]
- 38. Kanoh J., Ishikawa F.. spRap1 and spRif1, recruited to telomeres by taz1, are essential for telomere function in fission yeast. Curr. Biol. 2001; 11:1624–1630. [DOI] [PubMed] [Google Scholar]
- 39. Audry J., Maestroni L., Delagoutte E., Gauthier T., Nakamura T.M., Gachet Y., Saintomé C., Géli V., Coulon S.. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 2015; 34:1942–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J., Eisenstatt J.R., Audry J., Cornelius K., Shaughnessy M., Berkner K.L., Runge K.W.. A heterochromatin domain forms gradually at a new telomere and is dynamic at stable telomeres. Mol. Cell. Biol. 2018; 38:2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coulon S., Ramasubramanyan S., Alies C., Philippin G., Lehmann A., Fuchs R.P.. Rad8Rad5/Mms2-Ubc13 ubiquitin ligase complex controls translesion synthesis in fission yeast. EMBO J. 2010; 29:2048–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Audry J., Runge K.. Ccq1 restrains Mre11-mediated degradation of short telomeres. 2022; Research Square doi:24 March 2022, preprint: not peer reviewed 10.21203/rs.3.rs-1456395/v1. [DOI] [PMC free article] [PubMed]
- 43. Yanagida M. Clearing the way for mitosis: is cohesin a target?. Nat. Rev. Mol. Cell Biol. 2009; 10:489–496. [DOI] [PubMed] [Google Scholar]
- 44. Ferreira M.G. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004; 18:2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mochida S., Yanagida M.. Distinct modes of DNA damage response in s. pombe G0 and vegetative cells. Genes Cells. 2005; 11:13–27. [DOI] [PubMed] [Google Scholar]
- 46. Ferreira M.G., Cooper J.P.. The fission yeast taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell. 2001; 7:55–63. [DOI] [PubMed] [Google Scholar]
- 47. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moser B.A., Subramanian L., Khair L., Chang Y.-T., Nakamura T.M.. Fission yeast tel1(atm) and rad3(atr) promote telomere protection and telomerase recruitment. PLoS Genet. 2009; 5:e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naito T., Matsuura A., Ishikawa F.. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 1998; 20:203–206. [DOI] [PubMed] [Google Scholar]
- 50. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 51. Ma M., Rodriguez A., Sugimoto K.. Activation of ATR-related protein kinase upon DNA damage recognition. Curr. Genet. 2020; 66:327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Limbo O., Chahwan C., Yamada Y., Bruin R.A.M.de, Wittenberg C., Russell P.. Ctp1 is a cell-cycle-regulated protein that functions with mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007; 28:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Langerak P., Mejia-Ramirez E., Limbo O., Russell P.. Release of ku and MRN from DNA ends by mre11 nuclease activity and ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011; 7:e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fujita I., Tanaka M., Kanoh J.. Identification of the functional domains of the telomere protein rap1 in schizosaccharomyces pombe. PLoS One. 2012; 7:e49151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jun H.-I., Liu J., Jeong H., Kim J.-K., Qiao F.. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via ccq1. Genes Dev. 2013; 27:1917–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Escandell J.M., Carvalho E.S., Gallo-Fernández M., Reis C.C., Matmati S., Luís I.M., Abreu I.A., Coulon S., Ferreira M.G.. Ssu72 phosphatase is a conserved telomere replication terminator. EMBO J. 2019; 38:e100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matmati S., Vaurs M., Escandell J.M., Maestroni L., Nakamura T.M., Ferreira M.G., Géli V., Coulon S.. The fission yeast stn1-ten1 complex limits telomerase activity via its SUMO-interacting motif and promotes telomeres replication. Sci. Adv. 2018; 4:eaar2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garg M., Gurung R.L., Mansoubi S., Ahmed J.O., Davé A., Watts F.Z., Bianchi A.. Tpz1TPP1 SUMOylation reveals evolutionary conservation of SUMO-dependent stn1 telomere association. EMBO Rep. 2014; 15:871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miyagawa K., Low R.S., Santosa V., Tsuji H., Moser B.A., Fujisawa S., Harland J.L., Raguimova O.N., Go A., Ueno M.et al.. SUMOylation regulates telomere length by targeting the shelterin subunit tpz1(tpp1) to modulate shelterin-Stn1 interaction in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5950–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., Masai H.. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012; 26:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanoh Y., Matsumoto S., Fukatsu R., Kakusho N., Kono N., Renard-Guillet C., Masuda K., Iida K., Nagasawa K., Shirahige K.et al.. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 2015; 22:889–897. [DOI] [PubMed] [Google Scholar]
- 62. Kedziora S., Gali V.K., Wilson R.H.C., Clark K.R.M., Nieduszynski C.A., Hiraga S., Donaldson A.D.. Rif1 acts through protein phosphatase 1 but independent of replication timing to suppress telomere extension in budding yeast. Nucleic Acids Res. 2018; 46:3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Davé A., Cooley C., Garg M., Bianchi A.. Protein phosphatase 1 recruitment by rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014; 7:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hoerr R.E., Ngo K., Friedman K.L.. When the ends justify the means: regulation of telomere addition at double-strand breaks in yeast. Front. Cell Dev. Biol. 2021; 9:655377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aylon Y., Liefshitz B., Kupiec M.. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004; 23:4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dehé P.-M., Rog O., Ferreira M.G., Greenwood J., Cooper J.P.. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol. Cell. 2012; 46:797–808. [DOI] [PubMed] [Google Scholar]
- 67. Miller K.M., Ferreira M.G., Cooper J.P.. Taz1, rap1 and rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005; 24:3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Teixeira-Silva A., Saada A.A., Hardy J., Iraqui I., Nocente M.C., Fréon K., Lambert S.A.E.. The end-joining factor ku acts in the end-resection of double strand break-free arrested replication forks. Nat. Commun. 2017; 8:1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Matmati S., Lambert S.A.E., Géli V., Coulon S.. Telomerase repairs collapsed replication forks at telomeres. Cell Rep. 2020; 30:3312–3322. [DOI] [PubMed] [Google Scholar]
- 70. Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M.. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007; 445:506–510. [DOI] [PubMed] [Google Scholar]
- 71. Liu J., Hu X., Bao K., Kim J.-K., Zhang C., Jia S., Qiao F.. The cooperative assembly of shelterin bridge provides a kinetic gateway that controls telomere length homeostasis. Nucleic Acids Res. 2021; 49:8110–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang Y.-T., Moser B.A., Nakamura T.M.. Fission yeast shelterin regulates DNA polymerases and Rad3ATR kinase to limit telomere extension. PLoS Genet. 2013; 9:e1003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moser B.A., Subramanian L., Chang Y.-T., Noguchi C., Noguchi E., Nakamura T.M.. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009; 28:810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hiraga S., Monerawela C., Katou Y., Shaw S., Clark K.R., Shirahige K., Donaldson A.D.. Budding yeast rif1 binds to replication origins and protects DNA at blocked replication forks. EMBO Rep. 2018; 19:e46222–, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mattarocci S., Reinert J.K., Bunker R.D., Fontana G.A., Shi T., Klein D., Cavadini S., Faty M., Shyian M., Hafner L.et al.. Rif1 maintains telomeres and mediates DNA repair by encasing DNA ends. Nat. Struct. Mol. Biol. 2017; 24:588–595. [DOI] [PubMed] [Google Scholar]
- 76. Shubin C.B., Mayangsari R., Swett A.D., Greider C.W.. Rif1 regulates telomere length through conserved HEAT repeats. Nucleic Acids Res. 2021; 49:3967–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.