Abstract
Purpose
To compare temporal evolution of imaging features of coronavirus disease 2019 (COVID-19) and influenza in computed tomography and evaluate their predictive value for distinction.
Methods
In this retrospective, multicenter study 179 CT examinations of 52 COVID-19 and 44 influenza critically ill patients were included. Lung involvement, main pattern (ground glass opacity, crazy paving, consolidation) and additional lung and chest findings were evaluated by two independent observers. Additional findings and clinical data were compared patient-wise. A decision tree analysis was performed to identify imaging features with predictive value in distinguishing both entities.
Results
In contrast to influenza patients, lung involvement remains high in COVID-19 patients > 14 days after the diagnosis. The predominant pattern in COVID-19 evolves from ground glass at the beginning to consolidation in later disease. In influenza there is more consolidation at the beginning and overall less ground glass opacity (p = 0.002). Decision tree analysis yielded the following: Earlier in disease course, pleural effusion is a typical feature of influenza (p = 0.007) whereas ground glass opacities indicate COVID-19 (p = 0.04). In later disease, particularly more lung involvement (p < 0.001), but also less pleural (p = 0.005) and pericardial (p = 0.003) effusion favor COVID-19 over influenza. Regardless of time point, less lung involvement (p < 0.001), tree-in-bud (p = 0.002) and pericardial effusion (p = 0.01) make influenza more likely than COVID-19.
Conclusions
This study identified differences in temporal evolution of imaging features between COVID-19 and influenza. These findings may help to distinguish both diseases in critically ill patients when laboratory findings are delayed or inconclusive.
Abbreviations: COPD, Chronic obstructive pulmonary disease; COVID-19, Coronavirus disease 2019; CT, Computed tomography; HSCT, Haematopoietic stem cell transplantation; GGO, Ground glass opacity; HIV, Human immunodeficiency virus; ICC, Intraclass correlation coefficient; ICU, Intensive care unit; IQR, Interquartile range; PCR, Polymerase chain reaction; SD, Standard deviation; SOT, Solid organ transplantation
Keywords: COVID-19, Influenza, Pneumonia, Lung, Computed tomography
Highlights
-
•
Decision tree analysis helps to distinguish COVID-19 and Influenza.
-
•
Pleural effusion is a typical feature of influenza in early disease.
-
•
Ground glass opacities indicate COVID-19 in early disease.
-
•
Lung involvement remains high in COVID-19 patients > 14 days after the diagnosis.
-
•
Pleural and pericardial effusion favor influenza over COVID-19 in later disease.
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 is still challenging health-care systems due to new virus mutations with increased transmissibility and/or immune escape [1]. Compared to seasonal influenza, COVID-19 patients require longer hospitalization and have a higher complication rate [2].
In COVID-19, previous cross-sectional studies reported more round opacities [3], [4] and a larger extent of pathological lung involvement [3], less consolidation [5] and more ground glass opacity (GGO) [6], [7] as well as interlobular septal thickening [4], subpleural sparing and subpleural bands [6].
In influenza, however, pleural effusion [3], [4], [6], nodules and tree-in-bud opacities were reported more commonly by some studies [4], [5], whereas another study did not observe differences regarding tree-in-bud [8]. Consolidation [3], bronchial wall thickening and inferior lobe predominance [8] were also observed.
Distribution within the lung was reported as a feature to help differentiate between COVID-19 and influenza: Peripheral distribution is associated with COVID-19 [7], [8].
Since COVID-19 will not disappear despite availability of effective vaccines, and eventually will show the same seasonal peak as observed with influenza, it is of utmost clinical importance to distinguish between these two potentially fatal respiratory viral infections.
On imaging, COVID-19 lung abnormalities follow a typical course and tend to start with GGO followed by a crazy paving pattern and consolidation [9], [10], [11], typically peaking at day 9–13 after symptom onset [9], [11]. However, to the best of our knowledge, a longitudinal comparison between COVID-19 and influenza has not been done yet.
In this study, we compared clinical and imaging findings of critically ill patients infected with COVID-19 and influenza. Unlike previous studies, the temporal evolution of lung involvement and predominant lung pattern during the disease was assessed. Besides prognostic factors, diagnostic criteria, and their value for distinction of both conditions were evaluated.
2. Materials and methods
Ethics approval and informed consent was obtained for this multicenter study (EKOS 2018–01994, 2019–02173 and EKOS 2020–00908). In this retrospective cohort study, critically ill patients with COVID-19, hospitalized between March 2020 and March 2021 at St. Gallen Cantonal Hospital, Switzerland were compared to critically ill influenza patients from 2017/2018 and 2019/2020, hospitalized at six large Swiss Hospitals, that is St. Gallen Cantonal Hospital, Zurich University Hospital, Bern University Hospital, Basel University Hospital and Lausanne University HospitalAarau Cantonal Hospital.
All included patients required high-level of care (ICU or a specialized pulmonary unit). Patients with COVID-19 were enrolled in a prospective multicenter study (CRiPSI: COVID-19 Risk Prediction in Swiss ICUs-Trial) and required SARS-CoV-2 confirmed by rt-PCR or antigen assay. Influenza infections were laboratory-confirmed by PCR or antigen testing.
For both patient groups the following inclusion criteria were applied: informed consent and age ≥ 18 years. Patients were excluded if they declined to participate in the study, or if COVID-19 or influenza were only diagnosed after ICU discharge or if no CT examination was available.
2.1. Imaging
CT imaging protocols among the different centers were not strictly standardized, but imaging adhered to recommendations as follows: Patients were scanned in prone position, intubated patients were scanned in supine position. If CT angiography was required to evaluate for pulmonary embolism, patients were scanned in supine position with administration of intravenous iodinated contrast agent. Scanner settings were 100 kV, 50–100 mAs, depending on requirement for contrast agent or availability of automated dose modulation (e. g., automated tube voltage selection and automated tube current modulation). Reconstructions were made using iterative algorithms. Slice thickness was 5 mm or 2 mm for transverse and 5 mm for sagittal reconstructions. Imaging was performed as clinically indicated according to the referring physicians.
2.2. Image evaluation
Two radiologists (TF and YEB) with seven and eight years of experience in chest radiology, respectively, independently evaluated all CT examinations (axial and sagittal reconstructions), blinded for examination date, all clinical and microbiological information. Disagreements between both readers were processed as follows: Continuous variables: values were averaged; categorial variables: solved by consensus reading.
2.3. Evaluation of lobe involvement
Estimation of affected lung was performed as previously reported [11], [12] and was done lobe-wise (right upper lobe, middle lobe, right lower lobe, left upper lobe and left lower lobe) in percent (%) in each CT examination. Weighted mean of total lung involvement was calculated by use of previously described proportions [13].
2.4. Evaluation of main pattern
For each CT examination, the predominant pattern was chosen between the three following CT features: GGO (region of increased lung attenuation with vessels still visible), crazy paving (appearance of GGO with superimposed interlobular and intralobular septal thickening) or consolidation (homogeneous increased lung opacity with vessels not visible).
2.5. Evaluation of additional findings
Additional findings were evaluated as present or absent in each CT examination and were defined based on the most typical COVID-19 imaging features [14] and previously defined typical imaging features of influenza [3], [4], [5], [6], [7], [8], [15], [16]. Additional findings were: subpleural linear opacity, septal thickening, subpleural reticulation, air bronchogram, pleural thickening, bronchiectasis, bronchial wall thickening, tree-in-bud, pulmonary nodules, and vascular enlargement. Intrathoracic lymph node size (short-axis diameter), pleural effusion (if bilateral, both sides were summed up) and pericardial effusion were measured in millimeters. For evaluation of bronchial wall thickness (lobar, segmental and subsegmental), the T/D ratio (wall thickness (T) divided by total diameter of bronchus (D)) was measured. Values between 0.1 and 0.2 have been described as normal [17], [18]. In this study, a value above 0.2 was defined as bronchial wall thickening.
Nonspecific findings (neither systematically reported for COVID-19 nor influenza) were cavitation, halo sign, reverse halo sign and tracheal wall irregularities, that could reflect ulcers or pseudomembranes.
2.6. Clinical features
The following co-morbid features were recorded: Obesity, substance abuse, chronic obstructive pulmonary disease (COPD), asthma, neoplasm, hematological disease, hematopoietic stem cell transplantation (HSCT), solid organ transplantation (SOT), human immunodeficiency virus (HIV) infection, chronic renal failure, diabetes, cardiovascular disease, cerebrovascular disease, and pregnancy. The following medication prior to COVID-19 or influenza disease were recorded: Chronic steroid treatment (≥0.1 mg/kg/day prednisone equivalent) and immunosuppressive drugs. The following medication during the COVID-19 or influenza disease were recorded: Steroids and specific antiviral drugs. Additionally, bacterial respiratory co-infections were recorded according to assessment by infectious disease consultants and evaluated. The intubation status during the hospital stay was reviewed.
2.7. Data transformations
“Main pattern” was dichotomized into the three binary variables “consolidation”, “crazy paving”, and “ground glass opacity” being either present or absent. To simplify matters, multiple examinations per patients were reduced to one value for patients-wise comparisons. In case of involvement that was analyzed as a function of time, the method of least squares was used to compute the intercept and slope for each patient with more than one CT and the predicted involvement was calculated at the patient’s mean day after diagnosis. For all other variables, the mean was calculated for continuous variables and the maximum for binary variables (that were coded as 0 = absent and 1 = present) for each patient.
2.8. Statistics
Continuous variables with normal distribution were compared with a Student’s t-test for independent samples. Other continuous variables were compared with a Wilcoxon rank-sum test (with continuity correction) between COVID-19 and influenza patients. Categorical variables were compared with a Fisher’s exact test, intubation rates were compared with the chi-squared-test.
The intra-class correlation coefficient (ICC) was computed to assess the agreement between two radiologists rating the involvement of upper, lower, and total lung, left and right pleural effusion, lymph node size, and pericardial effusion in each of the 179 images. The ICC (two-way random-effects model for single rating) is reported. ICC is interpreted as poor for values below 0.50, as moderate for values between 0.50 and 0.75, as good for values between 0.75 and 0.90, and as excellent for values above 0.90 [19].
Cohen’s κ was computed to assess the agreement between the two radiologists rating various features as being either present or absent. The following classifications has been suggested to interpret the strength of the agreement based on the Cohen’s κ value [20]: Values > 0.4 were considered moderate, > 0.6 substantial and > 0.8 almost perfect agreement.
The association of diagnosis (influenza vs COVID-19) with combinations of involvement, GGO, air bronchogram, bronchiectasis, tree-in-bud, pulmonary nodules, pleural effusion, and pericardial effusion was further analyzed with tree-based models, which is a non-parametric procedure that works by partitioning the patients into subsets with similar response values using a set of splitting rules. The general procedure is as follows: Starting with the entire data set, a split is made at a specific cut point on one of the predictors chosen to maximize the difference in response between the resulting subsets. Each of the two subsets can be further subdivided using different predictors and/or different cut points. Splitting is associated with a test of significance for the difference in response between the two newly created subsets. Splitting continues as long as the resulting subsets are significantly different at the 5 % level (with or without correction for multiple testing). Analyses were mostly performed without correction for multiple testing and must therefore be regarded as exploratory.
2.9. Software
Most analyses were performed in the R programming language (version 4.0.2) (R Core Team, 2020). The package “psych” [21] was used to calculate ICC and Cohen’s κ. The package “tableone” [22] was used for descriptive statistics. The package “ggplot2″ [23] was used to plot the figures. The package “party” [24] was used to run the classification tree analysis. Comparison of lung involvement, main pattern and additional chest findings for COVID-19 patients and influenza patients with and without bacterial superinfection was done with Kruskal-Wallis Test (continuous variables) and Fisher’s exact test (categorical variables). Comparison of age was done with SPSS v. 25.0.0.2 (IBM Corp, Armok, NY) using Student’s t-test for independent samples.
3. Results
A total of 54 COVID-19 patients and 83 influenza patients were potentially eligible. None of the enrolled patients had SARS-CoV-2 and influenza coinfections. Two COVID-19 patients and 39 influenza patients had no CT examination, resulting in a total of 96 patients (52 COVID-19 and 44 influenza patients) with 179 CT examinations (26/179 unenhanced). Nearly half of all patients (47/96) had only one CT examination, and the remainder had 2–6 CTs per patient. The CTs were acquired between 7 days before and 62 days after the diagnosis.
3.1. Baseline characteristics
Patients’ mean age was 60.8 years (standard deviation (SD) 13.5 years, range 18.4–81.9 years), (COVID-19 patients 63.5 ± 9.5 years, range 35.2–80.7 years, influenza patients 57.6 ± 16.6 years, range 18.4–81.9 years). COVID-19 patients were significantly older (p = 0.04).
Overall, 34/96 (35.4 %), (12/52 (23.1 %) of COVID-19 patients and 22/44 (50.0 %) of influenza patients) were female. Males were more prevalent in the COVID-19 group (p = 0.01). Mortality was 21/52 (40.4 %) in COVID-19 patients and 10/44 (22.7 %) in influenza patients (p = 0.08). In the COVID-19 group, median time from disease specific symptom onset to the diagnosis was 3 days (IQR: 1–6 days), range − 3 to 14 days. In the influenza group, median time from disease specific symptom onset to the diagnosis was 4 days (IQR: 2–7 days), range 0–26 days.
3.2. Comparison of clinical data
Comparison of clinical data is given in Table 1. Obesity was more common among COVID-19 patients (p < 0.001), whereas substance abuse (p = 0.003), COPD (0.019) and chronic renal failure (0.022) were more common among influenza patients. Prior chronic treatment with steroids was more common among influenza patients (p = 0.046). COVID-19 patients were more commonly treated with steroids (p < 0.001), whereas influenza patients were more commonly treated with antivirals, which was oseltamivir in all cases (p < 0.001). Of patients with COVID-19, 23 (44 %) were intubated, and 19 (43 %) of patients with influenza were intubated, differences were not significant (p = 1).
Table 1.
Comparison of clinical data.
| Variable | COVID-19 | Influenza | p-value |
|---|---|---|---|
| N (number of patients) | 52 | 44 | |
| Clinical feature | |||
| Obesity (%) | 26 (50.0) | 4 (9.1) | < 0.001 |
| Substance abuse (%) | 0 (0.0) | 7 (15.9) | 0.003 |
| COPD (%) | 3 (5.8) | 10 (22.7) | 0.02 |
| Asthma (%) | 2 (3.8) | 0 (0.0) | 0.59 |
| Neoplasm (%) | 3 (5.8) | 7 (15.9) | 0.22 |
| Hematological disease (%) | 2 (3.8) | 6 (13.6) | 0.14 |
| HSCT (%) | 0 (0) | 0 (0) | NA |
| SOT (%) | 1 (1.9) | 1 (2.3) | 1.00 |
| HIV (%) | 0 (0) | 0 (0) | NA |
| Chronic renal failure (%) | 6 (11.5) | 14 (31.8) | 0.02 |
| Diabetes (%) | 20 (38.5) | 9 (20.5) | 0.08 |
| Cardiovascular disease (%) | 14 (26.9) | 14 (31.8) | 0.66 |
| Cerebrovascular disease (%) | 4 (7.7) | 4 (9.1) | 1.000 |
| Pregnancy (%) | 0 (0.0) | 1 (2.3) | 0.46 |
| Any other disease (%) | 25 (48.1) | 17 (38.6) | 0.41 |
| Prior immunosuppressive treatment | |||
| Steroids (%) | 4 (7.7) | 10 (22.7) | 0.046 |
| Immunosuppressive drug (%) | 2 (3.8) | 6 (13.6) | 0.14 |
| Treatment during disease | |||
| Steroids (%) | 51 (98.1) | 19 (43.2) | < 0.001 |
| Antivirals (%) | 7 (13.5) | 42 (95.5) | < 0.001 |
| Co-infection | |||
| Bacterial respiratory co-infections (%) | 28 (53.8) | 23 (52.3) | 1.000 |
COPD: Chronic obstructive pulmonary disease, HSCT: Hematopoietic stem cell transplantation, SOT: Solid organ transplant. Prior treatment with steroids: ≥ 0.1 mg/kg/day prednisone equivalent.
3.3. Interrater reliability
Interrater reliability was excellent for total lung, upper and lower lung involvements, each with ICC > 0.9. It was good for left and right pleural effusion as well as for pericardial effusion, and moderate for lymph node size. Cohen’s κ could not be computed for reverse halo sign, where for 178 images, both readers rated “absent” and for one image only, one reader rated “present”. For most other imaging features, the agreement was substantial to near perfect.
3.4. Comparison of lung involvement
There was greater lung involvement for COVID-19 patients with median differences of 21.5 % for total lung involvement (p < 0.001), of 20.6 % for upper lobe involvement (p < 0.001) and of 7.4 % for lower lobe involvement (p = 0.005) (Table 2).
Table 2.
Median involvement (IQR) of the total lung, the upper lobe and the lower lobe and presence of consolidation, crazy paving and ground glass as main pattern for COVID-19 and influenza patients.
| Variable | COVID-19 | Influenza | p-value |
|---|---|---|---|
| N (number of patients) | 52 | 44 | |
| Median involvement | |||
| Total lung involvement (%) (median [IQR]) | 65.8 [54.9, 82.9] | 44.3 [12.6, 59.6] | < 0.001 |
| Upper lobe involvement (%) (median [IQR]) | 32.1 [24.3, 39.1] | 11.5 [3.4, 25.4] | < 0.001 |
| Lower lobe involvement (%) (median [IQR]) | 37.0 [28.8, 42.3] | 29.6 [9.8, 37.8] | 0.005 |
| Presence of main pattern | |||
| Consolidation (%) | 16 (30.8) | 22 (50.0) | 0.06 |
| Crazy paving (%) | 21 (40.4) | 14 (31.8) | 0.40 |
| Ground glass opacity (%) | 37 (71.2) | 17 (38.6) | 0.002 |
3.5. Comparison of lung involvement over time
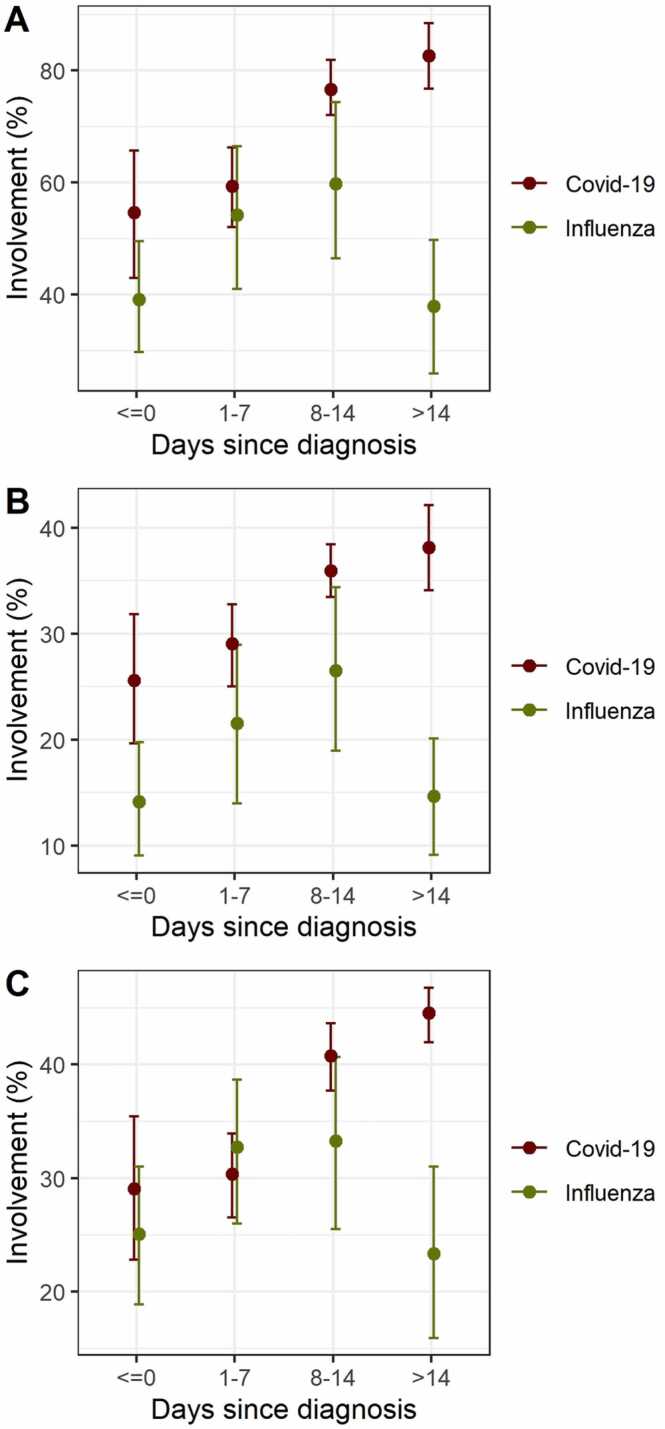
Time-dependent evaluation of lung involvement revealed a continuous increase over time for total lung, upper and lower lobe involvement in COVID-19 patients, whereas lung involvement increased in influenza patients until the end of the second week with a subsequent decrease (Fig. 1).
Fig. 1.
Mean degree of involvements of total lung (A), upper lobes (B), and lower lobes (C) in COVID-19 (red) and influenza patients (green) by time. Each time bin comprises only one (the first) measurement per patient due to the otherwise dependent nature of the data. Error bars show mean and bootstrapped 95 % confidence intervals for days − 7 to 0 (n = 17 and 23 for COVID-19 and influenza), days 1–7 (n = 33 and 18 for COVID-19 and influenza), days 8–14 (n = 23 and 11 for COVID-19 and influenza) and > 14 days after diagnosis (n = 20 and 14 for COVID-19 and influenza). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Comparison of main pattern
In an overall comparison of observed main patterns (consolidation, crazy paving and GGO), GGO was the predominant pattern in COVID-19 patients (Table 2). While GGO was the most common main pattern in COVID-19 patients (71.2 % vs. 38.6 % in influenza, p = 0.002), consolidation was the most common main pattern in patients with influenza (50.0 % vs. 30.8 % in COVID-19, p = 0.06). Presence of bacterial superinfection did not alter these differences between COVID-19 and influenza (data not shown).
3.7. Temporal comparison of predominant main pattern
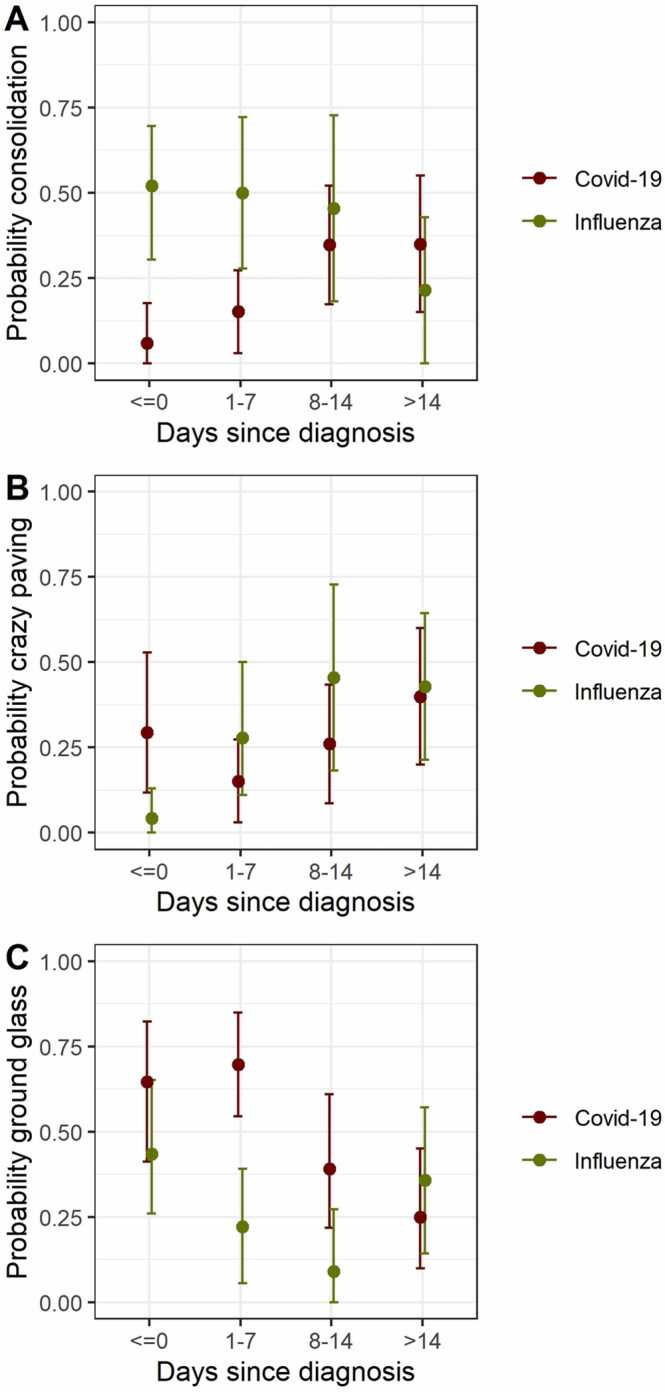
For the majority of COVID-19 cases, GGO was changing from present to absent or was not changing over time. Changes from absent to present later in the course of the disease were uncommon. For the majority of influenza patients, GGO was not changing over the whole period of observation. The probability of GGO was lower for influenza than for COVID-19 patients, especially in earlier disease (≤ day seven). The probability seemed to be declining over time for both patient groups (Fig. 2a). Although crazy paving could change within patients from present to absent and vice versa, for more than half of all patients the predominant crazy paving pattern did not change over time. Crazy paving as main pattern was slightly increasing over time both for COVID-19 and for influenza patients (Fig. 2b). The probability for consolidation as main pattern was increasing over time for COVID-19 patients and decreasing for influenza patients (Fig. 2c).
Fig. 2.
Predominant main pattern (ground glass opacity, A, crazy paving, B and consolidation, C) in COVIOD-19 (red) and influenza patients (green) by time. Each time bin comprises only one (the first) measurement per patient due to the otherwise dependent nature of the data. Error bars show mean and bootstrapped 95 % confidence intervals for days − 7 to 0 (n = 17 and 23 for COVID-19 and influenza), days 1–7 (n = 33 and 18 for COVID-19 and influenza), days 8–14 (n = 23 and 11 for COVID-19 and influenza) and > 14 days after diagnosis (n = 20 and 14 for COVID-19 and influenza). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.8. Comparison of additional findings
Comparison of additional findings between COVID-19 and influenza patients indicated that air bronchogram was present more often in COVID-19 (63.5 %) than influenza patients (38.6 %, p = 0.024, Table 3). Conversely, tree-in-bud (p < 0.001) and pulmonary nodules (p = 0.04) were present more often in influenza (40.9 % and 18.2 %, respectively) than COVID-19 patients (5.8 % and 3.8 %, respectively). Moreover, the size of pleural and pericardial effusion was larger in influenza than COVID-19 patients (Table 3). Taking the presence of bacterial superinfection into account slightly changed these results (Table 4). Air bronchogram and pleural effusion were not different any more (p = 0.08 and p = 0.08, respectively) while tracheal wall irregularities were present only in those without bacterial superinfection and more commonly in COVID-19 (16.7 % in COVID-19 patients vs. 4.8 % in influenza patients, p = 0.02).
Table 3.
Comparison of additional chest findings for COVID-19 and influenza patients.
| COVID-19 | Influenza | p-value | |
|---|---|---|---|
| N (number of patients) | 52 | 44 | |
| Subpleural linear opacity (%) | 13 (25.0) | 16 (36.4) | 0.27 |
| Septal thickening (%) | 34 (65.4) | 20 (45.5) | 0.06 |
| Subpleural reticulation (%) | 9 (17.3) | 11 (25.0) | 0.45 |
| Air bronchogram (%) | 33 (63.5) | 17 (38.6) | 0.02 |
| Pleural thickening (%) | 5 (9.6) | 0 (0.0) | 0.06 |
| Bronchiectasis (%) | 10 (19.2) | 2 (4.5) | 0.03 |
| Bronchial wall thickening (%) | 14 (26.9) | 12 (27.3) | 1.000 |
| Tree-in-bud (%) | 3 (5.8) | 18 (40.9) | < 0.001 |
| Pulmonary nodules (%) | 2 (3.8) | 8 (18.2) | 0.04 |
| Vascular enlargement (%) | 10 (19.2) | 5 (11.4) | 0.40 |
| Lymph node size (median [IQR]) | 7.9 [7.0, 9.8] | 8.5 [7.9, 9.1] | 0.33 |
| Pleural effusion (median [IQR]) | 0.0 [0.0, 8.1] | 7.5 [0.0, 19.6] | 0.02 |
| Pericardial effusion (median [IQR]) | 0.0 [0.0, 0.5] | 1.0 [0.5, 3.5] | < 0.001 |
| Cavitation (%) | 6 (11.5) | 1 (2.3) | 0.12 |
| Halo sign (%) | 7 (13.5) | 6 (13.6) | 1.00 |
| Reverse halo sign (%) | 0 (0.0) | 0 (0.0) | NA |
| Tracheal wall irregularity (%) | 4 (7.7) | 1 (2.3) | 0.37 |
Table 4.
Comparison of additional chest findings for COVID-19 and influenza patients with and without bacterial superinfection.
| COVID-19 with bacterial superinfection | COVID-19 without bacterial superinfection | Influenza with bacterial superinfection | Influenza without bacterial superinfection | p-value | |
|---|---|---|---|---|---|
| N (number of patients) | 28 | 24 | 23 | 21 | |
| Subpleural linear opacity (%) | 9 (32.1) | 4 (16.7) | 7 (30.4) | 9 (42.9) | 0.29 |
| Septal thickening (%) | 19 (67.9) | 15 (62.5) | 13 (56.5) | 7 (33.3) | 0.10 |
| Subpleural reticulation (%) | 6 (21.4) | 3 (12.5) | 7 (30.4) | 4 (19.0) | 0.52 |
| Air bronchogram (%) | 19 (67.9) | 14 (58.3) | 10 (43.5) | 7 (33.3) | 0.08 |
| Pleural thickening (%) | 3 (10.7) | 2 (8.3) | 0 (0.0) | 0 (0.0) | 0.22 |
| Bronchiectasis (%) | 8 (28.6) | 2 (8.3) | 2 (8.7) | 0 (0.0) | 0.02 |
| Bronchial wall thickening (%) | 9 (32.1) | 5 (20.8) | 6 (26.1) | 6 (28.6) | 0.86 |
| Tree-in-bud (%) | 2 (7.1) | 1 (4.2) | 11 (47.8) | 7 (33.3) | < 0.001 |
| Pulmonary nodules (%) | 0 (0.0) | 2 (8.3) | 4 (17.4) | 4 (19.0) | 0.06 |
| Vascular enlargement (%) | 7 (25.0) | 3 (12.5) | 1 (4.3) | 4 (19.0) | 0.21 |
| Lymph node size (median [IQR]) | 7.7 [6.9, 9.5] | 8.5 [7.0, 10.0] | 8.3 [7.8, 9.1] | 8.5 [8.0, 9.0] | 0.70 |
| Pleural effusion (median [IQR]) | 0.8 [0.0, 12.8] | 0.0 [0.0, 7.0] | 6.8 [0.0, 16.4] | 9.6 [0.0, 29.2] | 0.08 |
| Pericardial effusion (median [IQR]) | 0.0 [0.0, 0.5] | 0.0 [0.0, 0.6] | 1.0 [0.4, 2.5] | 1.5 [0.5, 4.0] | < 0.001 |
| Cavitation (%) | 5 (17.9) | 1 (4.2) | 1 (4.3) | 0 (0.0) | 0.10 |
| Halo sign (%) | 3 (10.7) | 4 (16.7) | 5 (21.7) | 1 (4.8) | 0.39 |
| Reverse halo sign (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| Tracheal wall irregularity (%) | 0 (0.0) | 4 (16.7) | 0 (0.0) | 1 (4.8) | 0.02 |
3.9. Outcome prediction
For influenza and COVID-19 patients, none of the evaluated variables seemed to be strongly associated with death (p-value >0.05 in bivariate analyses for all, including maximum lung involvement).
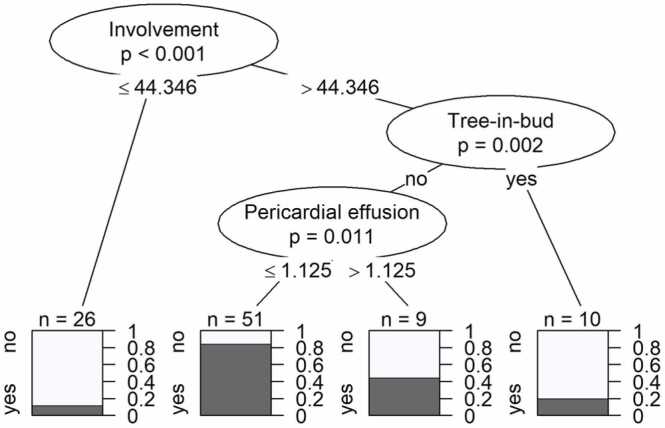
3.10. Classification tree analysis
A classification tree for prediction of COVID-19 was constructed for the entire observation time with Bonferroni correction for multiple testing (Fig. 3). Degree of lung involvement, absence of tree-in-bud, and smaller pericardial effusion were significant predictors for COVID-19 (compared to influenza).
Fig. 3.
Classification tree for the prediction of COVID-19 for entire observation time (with Bonferroni correction). The tree illustrates how the data set was split at specific cut points of one of the predictors. Resulting data subsets represent groups of patients with COVID-19 diagnosis (dark parts of bar plots). P-values at each split indicate the significance of the relationship between the predictor and COVID-19 diagnosis among the patients considered at this split. Splitting criteria are indicated on the branches. Involvement in percent (%) of total lung, pericardial effusion in millimeter (mm).
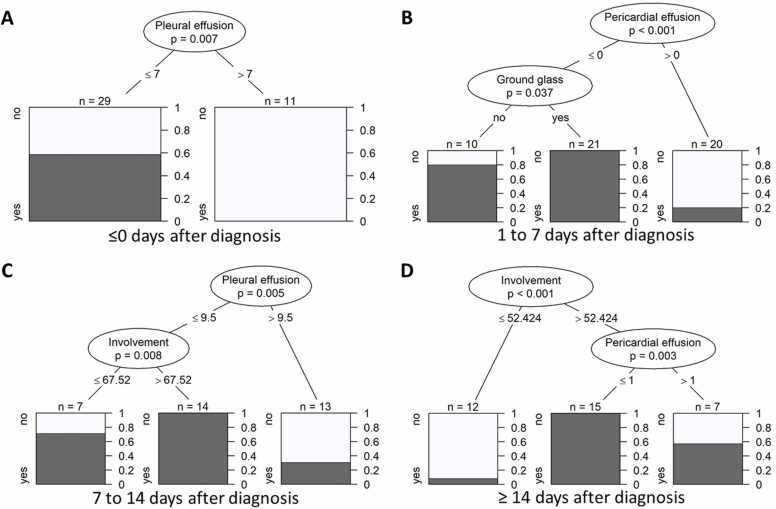
Four different classification trees for the prediction of COVID-19 were developed for different time points (Fig. 4a-d). For the first time bin, i.e., ≤ 0 days after diagnosis, only pleural effusion was retained: Larger volumes of pleural effusion were associated with influenza (Fig. 4a). For the second time bin, i.e., 1–7 days after diagnosis, pericardial effusion and GGO were retained in the partitioning: A lack of pericardial effusion at 1–7 days after diagnosis was a significant predictor for COVID-19, especially if in combination with GGO (Fig. 4b). For the third time bin, i.e., 8–14 days after diagnosis, pleural effusion and lung involvement were retained in the partitioning: Pleural effusion ≤ 9.5 cm (both sides summed up) at 8–14 days after diagnosis was a significant predictor for COVID-19, especially if in combination with larger total lung involvement (Fig. 4c). For the fourth time bin, i.e., > 14 days after diagnosis, pleural effusion and lung involvement were retained in the partitioning:Pleural effusion ≤ 9.5 cm (both sides summed up) at > 14 days after diagnosis was a significant predictor for COVID-19, especially if in combination with larger total lung involvement (Fig. 4d). Examples of imaging features associated with influenza are given in Fig. 5.
Fig. 4.
Classification tree for the prediction of COVID-19 for different time points: at ≤ 0 days after diagnosis (A), 0–7 days after diagnosis (B), 7–14 days after diagnosis (C) and ≥ 14 days after diagnosis (D) without Bonferroni correction. For each time bin, only one (the first) measurement per patient was included in the analysis. The trees illustrate how the data set was split at specific cut points of one of the predictors. Resulting data subsets represent groups of patients with COVID-19 diagnosis (dark parts of bar plots). P-values at each split indicate the significance of the relationship between the predictor and COVID-19 diagnosis among the patients considered at this split. Splitting criteria are indicated on the branches. Involvement in percent (%) of total lung, pericardial effusion in millimeter (mm), pleural effusion in millimeter (mm).
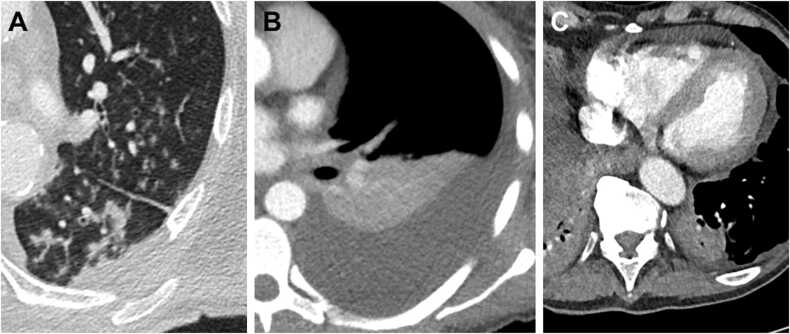
Fig. 5.
Examples of imaging features associated with influenza: Tree-in-bud (A) a in the left upper and lower lobe in a 60- years old male patient, one day after symptom onset at the day of the influenza diagnosis and at the day of ICU admission. Pleural effusion (B) in a 41- years old female patient 12 days after symptom onset, seven days after influenza diagnosis and six days after ICU admission. Pericardial effusion (C) in a 76- years old male patient, 27 days after symptom onset, 17 days after influenza diagnosis and 11 days after ICU admission.
4. Discussion
This is the first study that compares temporal evolution of imaging features of COVID-19 and influenza in critically ill patients. Unlike influenza, lung involvement remains high in COVID-19 > 14 days after the diagnosis. The predominant pattern in COVID-19 evolves from GGO at the beginning to consolidation in later disease. In contrast, the predominant pattern in influenza is more uniformly distributed, showing especially more consolidation at the beginning and less GGO overall.
According to our decision tree analysis, during the early stage of the disease larger volumes of pleural effusion and pericardial effusion favors the diagnosis of influenza over COVID-19 whereas ground glass opacities indicate COVID-19. At later stages of the disease, larger total lung involvement, but also less pleural and pericardial effusion favor COVID-19 over influenza. Regardless of the time point, a lower percentage of lung involvement, tree-in-bud, and pericardial effusion favor influenza over COVID-19.
While obesity is a well-known risk factor for both influenza and COVID-19, our study showed a higher rate of obese patients in the critically ill COVID-19 cohort. Underlying lung and kidney diseases have been more commonly observed among influenza patients [25], which supports the findings of our study. In general, patients with underlying disease tend to be more susceptible to viral infections and a severe disease course.
Overall lung involvement was higher in COVID-19 patients, which is supported by one study [3]. In a meta-analysis from 2020 including 33 studies and 1911 patients suffering from COVID-19 and influenza, no overlapping imaging features were identified, except for a higher prevalence of peripheral distribution and involvement of upper and middle lobes in COVID-19 [15]. This was also stated in a review article published in 2020, including 17 studies [16]. Greater lung involvement is probably the imaging surrogate for a longer ICU requirement and intubation time [26] and serves as an important predictor for COVID-19 on imaging. Intubation can lead to ventilation induced lung injury [27] which may be difficult to distinguish from COVID-19 or influenza related opacity. While intubation rates were similar in both groups, we did not have detailed information on ventilator settings which might have affected ventilation-induced lung injury.
Consistent with previous reports, consolidation was found more often in influenza compared to COVID-19 [3,5] and GGO more frequently in COVID-19 [6,7]. Temporal evaluation revealed a difference especially at the beginning of the disease between COVID-19 and influenza and presence of GGO serves as a predictor for COVID-19. In this study pleural effusion served as a time-dependent predictor favoring influenza, which is in line with previous results. Indeed, pleural effusion is more common in influenza [3,4,6] but uncommon in COVID-19 [28]. In this study, regardless of time point, tree-in-bud was a predictor of influenza. One previous study reported tree-in-bud as imaging finding of influenza [4], another study found no difference [8]. Additionally, our results indicate that pericardial effusion is one of the most powerful predictors for influenza. Pericardial effusion is uncommon in COVID-19 but has been reported in influenza patients [29], [30], [31], [32].
Sensitivity and specificity for distinguishing COVID-19 and influenza based on CT features have been reported to be 0.87–0.90 and 0.66–0.96 [7], [33] but interobserver agreement between radiologists seems to be rather low (kappa 0.41–0.59) [6]. Computer-based algorithms have shown to achieve an equal sensitivity and specificity of 0.62–0.90 and 0.88–0.91, respectively [5,34].
While COVID-19 and influenza molecular tests from respiratory samples are considered robust and gold standard in establishing a correct diagnosis, they can be hampered by availability, long turn-around times and have considerably lower sensitivity when applied to upper respiratory samples when patients present late [35]. In contrast, rapid antigen testing has reduced sensitivity compared to molecular diagnostics in both COVID-19 and influenza [36], [37]. As evident from this cohort, there might be delays of clinical presentation which might affect laboratory assays and point towards a potential value of readily available CT results in diagnosing and distinguishing both diseases.
The study has the following strengths: A well characterized population of critically ill patients was evaluated with up to six CT examinations per patient throughout the course of severe COVID-19 or influenza. This is an increasingly relevant clinical distinction as we approach endemicity of COVID-19, which involves a greatly different management strategy compared to severe influenza. Importantly, and in contrast to most previous studies, our study took the time-dependent nature of many imaging features into account. Our decision tree analysis resulted in easy to use and clearly distinctive time-dependent prediction algorithms.
The study has the following limitations: A major drawback is the relatively small sample size as well as the retrospective design of the study. Time and frequency of CT examinations were very heterogeneous in our patient cohort, since performed at different hospitals and according to the clinical necessity with fewer images at later time points. Thus, our results should be confirmed in prospective studies. Moreover, as multiple testing was not corrected for in the 4 different time-bin specific classification trees, those results should be regarded as exploratory. COVID-19 patients were significantly older compared to influenza patients which might have affected susceptibility to lung injury either through virus-induced changes or due to comorbidities. Finally, since patients from the first COVID-19 wave in Switzerland were included in this study, imaging characteristics may vary with other, different COVID-19 variants we have observed since then. However, recent data do not suggest relevant CT differences between COVID-19 variants [38].
5. Conclusion
In conclusion, this study shows a greater total lung involvement in critically ill COVID-19 patients compared to influenza patients throughout their clinical course. Main patterns differed between COVID-19 and influenza patients over time. We provide a clearly, distinctive, and time-dependent decision tree analysis which is easy to use and may help to distinguish both conditions in critically ill patients when laboratory findings are delayed or inconclusive.
Funding
The study was in part funded through the Cantonal Hospital St. Gallen (Forschungsfond), the Swiss Network on Fungal Diseases (FUNGINOS) and internal funds of the Division of Infectious Diseases & Hospital Epidemiology and of the Department of Radiology.
Ethical approval details
Approving body: Eastern Switzerland Ethics Committee, regestration number 2020-00908 and 2019-02173. Informed consent was obtained from all individuals, participating in this study.
CRediT authorship contribution statement
Tim Fischer: Conceptualization, Data curation, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Yassir El Baz: Investigation, Validation, Writing – review & editing. Giulia Scanferla: Investigation, Validation, Writing – review & editing. Nicole Graf: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – review & editing. Frederike Waldeck: Investigation, Validation, Writing – review & editing. Gian-Reto Kleger: Investigation, Validation, Writing – review & editing. Thomas Frauenfelder: Conceptualization, Data curation, Investigation, Methodology, Resources, Software, Validation, Writing – original draft, Writing – review & editing. Jens Bremerich: Investigation, Validation, Writing – review & editing. Sabine Schmidt Kobbe: Investigation, Validation, Writing – review & editing. Jean-Luc Pagani: Investigation, Validation, Writing – review & editing. Sebastian Schindera: Investigation, Validation, Writing – review & editing, Writing – review & editing. Anna Conen: Investigation, Validation, Writing – review & editing. Simon Wildermuth: Investigation, Validation, Writing – review & editing. Sebastian Leschka: Investigation, Validation, Writing – review & editing. Carol Strahm: Investigation, Validation, Writing – review & editing. Stephan Waelti: Investigation, Validation, Writing – review & editing. Tobias Johannes Dietrich: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. Werner C. Albrich: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. All authors have read and agreed to the published version of the manuscript.
Conflicting interests
The authors declare that they have no conflict of interests.
Contributor Information
Tim Fischer, Email: tim.fischer@kssg.ch.
Yassir El Baz, Email: yassir.elbaz@kssg.ch.
Giulia Scanferla, Email: giulia.scanferla@kssg.ch.
Nicole Graf, Email: nicole.graf@kssg.ch.
Frederike Waldeck, Email: frederike.waldeck@uksh.de.
Gian-Reto Kleger, Email: gian-reto.kleger@kssg.ch.
Thomas Frauenfelder, Email: thomas.frauenfelder@usz.ch.
Jens Bremerich, Email: jens.bremerich@usb.ch.
Sabine Schmidt Kobbe, Email: sabine.schmidt@chuv.ch.
Jean-Luc Pagani, Email: jean-luc.pagani@chuv.ch.
Sebastian Schindera, Email: sebastian.schindera@ksa.ch.
Anna Conen, Email: anna.conen@ksa.ch.
Simon Wildermuth, Email: simon.wildermuth@kssg.ch.
Sebastian Leschka, Email: sebastian.leschka@kssg.ch.
Carol Strahm, Email: carol.strahm@kssg.ch.
Stephan Waelti, Email: stephan.waelti@kssg.ch.
Tobias Johannes Dietrich, Email: tobias.dietrich@kssg.ch.
Werner C. Albrich, Email: werner.albrich@kssg.ch.
Data Availability
Data available on request from the corresponding author.
References
- 1.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2-what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 2.Deng L.-S., Yuan J., Ding L., Chen Y.-L., Zhao C.-H., Chen G.-Q., Li X.-H., Li X.-H., Luo W.-T., Lan J.-F., Tan G.-Y., Tang S.-H., Xia J.-Y., Liu X. Comparison of patients hospitalized with COVID-19, H7N9 and H1N1. Infect. Dis. Poverty. 2020;9:163. doi: 10.1186/s40249-020-00781-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S.-H., Wi Y.M., Lim S., Han K.-T., Bae I.-G. Differences in clinical characteristics and chest images between coronavirus disease 2019 and influenza-associated pneumonia. Diagnostic. 2021;11:261. doi: 10.3390/diagnostics11020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M., Zeng W., Wen Y., Zheng Y., Lv F., Xiao K. COVID-19 pneumonia: CT findings of 122 patients and differentiation from influenza pneumonia. Eur. Radiol. 2020;30:5463–5469. doi: 10.1007/s00330-020-06928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou M., Yang D., Chen Y., Xu Y., Xu J.-F., Jie Z., Yao W., Jin X., Pan Z., Tan J., Wang L., Xia Y., Zou L., Xu X., Wei J., Guan M., Yan F., Feng J., Zhang H., Qu J. Deep learning for differentiating novel coronavirus pneumonia and influenza pneumonia. Ann. Transl. Med. 2021;9:111. doi: 10.21037/atm-20-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarei F., Jalli R., Iranpour P., Sefidbakht S., Soltanabadi S., Rezaee M., Jahankhah R., Manafi A. Differentiation of chest CT findings between influenza pneumonia and COVID-19: interobserver agreement between radiologists. Acad. Radiol. 2021;28:1331–1338. doi: 10.1016/j.acra.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao S., Huang Z., Zeng H., Chen Z., Luo F., Zhang C., Song B. Combining initial chest CT with clinical variables in differentiating coronavirus disease 2019 (COVID-19) pneumonia from influenza pneumonia. Sci. Rep. 2021;11:6422. doi: 10.1038/s41598-021-85779-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Wei R., Rao G., Zhu J., Song B. Characteristic CT findings distinguishing 2019 novel coronavirus disease (COVID-19) from influenza pneumonia. Eur. Radiol. 2020;30:4910–4917. doi: 10.1007/s00330-020-06880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwee T.C., Kwee R.M. Chest CT in COVID-19: what the radiologist needs to know. Radiographics. 2020;40:1848–1865. doi: 10.1148/rg.2020200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X., Shi H., Zhou M. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020;296:55–64. doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li K., Fang Y., Li W., Pan C., Qin P., Zhong Y., Liu X., Huang M., Liao Y., Li S. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur. Radiol. 2020;30:4407–4416. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada Y., Yamada M., Yokoyama Y., Tanabe A., Matsuoka S., Niijima Y., Narita K., Nakahara T., Murata M., Fukunaga K., Chubachi S., Jinzaki M. Differences in lung and lobe volumes between supine and standing positions scanned with conventional and newly developed 320-detector-row upright CT: intra-individual comparison. RES. 2020;99:598–605. doi: 10.1159/000507265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Lan Y., Yao X., Lin S., Xie B. The chest CT features of coronavirus disease 2019 (COVID-19) in China: a meta-analysis of 19 retrospective studies. Virol. J. 2020;17:159. doi: 10.1186/s12985-020-01432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altmayer S., Zanon M., Pacini G.S., Watte G., Barros M.C., Mohammed T.-L., Verma N., Marchiori E., Hochhegger B. Comparison of the computed tomography findings in COVID-19 and other viral pneumonia in immunocompetent adults: a systematic review and meta-analysis. Eur. Radiol. 2020;30:6485–6496. doi: 10.1007/s00330-020-07018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onigbinde S.O., Ojo A.S., Fleary L., Hage R. Chest computed tomography findings in COVID-19 and influenza: a narrative review. Biomed. Res. Int. 2020;2020:6928368. doi: 10.1155/2020/6928368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka S., Uchiyama K., Shima H., Ueno N., Oish S., Nojiri Y. Bronchoarterial ratio and bronchial wall thickness on high-resolution CT in asymptomatic subjects: correlation with age and smoking. AJR Am. J. Roentgenol. 2003;180:513–518. doi: 10.2214/ajr.180.2.1800513. [DOI] [PubMed] [Google Scholar]
- 18.Elicker B., Webb W. 1st ed. Lippincott Williams & Wilkins; Philadelphia: 2013. Fundamentals of High-Resolution Lung CT: Common Findings, Common Patterns, Common Diseases, and Differential Diagnosis. [Google Scholar]
- 19.Koo T.K., Li M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 21.W. Revelle, psych: Procedures for Psychological, Psychometric, and Personality Research, 2021. 〈https://CRAN.R-project.org/package=psych〉 (Accessed 3 February 2022).
- 22.K. Yoshida, A. Bartel, Create “Table 1″ to Describe Baseline Characteristics with or without Propensity Score Weights [R package tableone version 0.12.0], 2020. 〈https://CRAN.R-project.org/package=tableone〉 (Accessed 11 May 2021).
- 23.Wickham H., Grolemund G. O’Reilly Media, Inc; 2016. R for Data Science: Import, Tidy, Transform, Visualize, and Model Data. [Google Scholar]
- 24.Hothorn T., Hornik K., Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J. Comput. Graph. Stat. 2006;15:651–674. doi: 10.1198/106186006×133933. [DOI] [Google Scholar]
- 25.Shen C., Tan M., Song X., Zhang G., Liang J., Yu H., Wang C. Comparative analysis of early-stage clinical features between COVID-19 and influenza A H1N1 virus pneumonia. Front. Public Health. 2020;8:206. doi: 10.3389/fpubh.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G., Bonanomi E., Cabrini L., Carlesso E., Castelli G., Cattaneo S., Cereda D., Colombo S., Coluccello A., Crescini G., Forastieri Molinari A., Foti G., Fumagalli R., Iotti G.A., Langer T., Latronico N., Lorini F.L., Mojoli F., Natalini G., Pessina C.M., Ranieri V.M., Rech R., Scudeller L., Rosano A., Storti E., Thompson B.T., Tirani M., Villani P.G., Pesenti A., Cecconi M. Risk factors associated with mortality among patients with COVID-19 in intensive care units in Lombardy, Italy. JAMA Intern. Med. 2020;180:1–11. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cressoni M., Gotti M., Chiurazzi C., Massari D., Algieri I., Amini M., Cammaroto A., Brioni M., Montaruli C., Nikolla K., Guanziroli M., Dondossola D., Gatti S., Valerio V., Vergani G.L., Pugni P., Cadringher P., Gagliano N., Gattinoni L. Mechanical power and development of ventilator-induced lung injury. Anesthesiology. 2016;124:1100–1108. doi: 10.1097/ALN.0000000000001056. [DOI] [PubMed] [Google Scholar]
- 28.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am. J. Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 29.Martín-Lázaro J.F., Homs C., Benito R., Pedro A.S., Suárez M.A. Chronic pericardial effusion secondary to a influenza virus A (H1N1)/2009 infection. Turk. Kardiyol. Dern. Ars. 2013;41:157–160. doi: 10.5543/tkda.2013.18827. [DOI] [PubMed] [Google Scholar]
- 30.Spoto S., Valeriani E., Locorriere L., Anguissola G.B., Pantano A.L., Terracciani F., Riva E., Ciccozzi M., Costantino S., Angeletti S. Influenza B virus infection complicated by life-threatening pericarditis: a unique case-report and literature review. BMC Infect. Dis. 2019;19:40. doi: 10.1186/s12879-018-3606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koranyi K., Yontz D., Rohrer Z., Leber A., Ramilo O. Pericardial effusion complicating novel influenza A (H1N1) infection in an infant. Pediatr. Infect. Dis. J. 2010;29:782–783. doi: 10.1097/INF.0b013e3181de4952. [DOI] [PubMed] [Google Scholar]
- 32.Vergara-Uzcategui C.E., López-Rondón É., Cabrera R. Pericardial effusion in a pediatric patient with influenza A H3N2. Arch. Cardiol. Mex. 2019;89:412–414. doi: 10.24875/ACM.19000052. [DOI] [PubMed] [Google Scholar]
- 33.Fink N., Rueckel J., Kaestle S., Schwarze V., Gresser E., Hoppe B., Rudolph J., Goller S., Kunz W.G., Ricke J., Sabel B.O. Evaluation of patients with respiratory infections during the first pandemic wave in Germany: characteristics of COVID-19 versus non-COVID-19 patients. BMC Infect. Dis. 2021;21:167. doi: 10.1186/s12879-021-05829-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y., Zhang Z., Liu S., Li X., Yang Y., Ma J., Li Z., Zhou J., Jiang Y., He B. CT-based radiomics combined with signs: a valuable tool to help radiologist discriminate COVID-19 and influenza pneumonia. BMC Med. Imaging. 2021;21:31. doi: 10.1186/s12880-021-00564-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller T.E., Garcia Beltran W.F., Bard A.Z., Gogakos T., Anahtar M.N., Astudillo M.G., Yang D., Thierauf J., Fisch A.S., Mahowald G.K., Fitzpatrick M.J., Nardi V., Feldman J., Hauser B.M., Caradonna T.M., Marble H.D., Ritterhouse L.L., Turbett S.E., Batten J., Georgantas N.Z., Alter G., Schmidt A.G., Harris J.B., Gelfand J.A., Poznansky M.C., Bernstein B.E., Louis D.N., Dighe A., Charles R.C., Ryan E.T., Branda J.A., Pierce V.M., Murali M.R., Iafrate A.J., Rosenberg E.S., Lennerz J.K. Clinical sensitivity and interpretation of PCR and serological COVID-19 diagnostics for patients presenting to the hospital. FASEB J. 2020;34:13877–13884. doi: 10.1096/fj.202001700RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu V.T., Schwartz N.G., Donnelly M.A.P., Chuey M.R., Soto R., Yousaf A.R., Schmitt-Matzen E.N., Sleweon S., Ruffin J., Thornburg N., Harcourt J.L., Tamin A., Kim G., Folster J.M., Hughes L.J., Tong S., Stringer G., Albanese B.A., Totten S.E., Hudziec M.M., Matzinger S.R., Dietrich E.A., Sheldon S.W., Stous S., McDonald E.C., Austin B., Beatty M.E., Staples J.E., Killerby M.E., Hsu C.H., Tate J.E., Kirking H.L., Matanock A. COVID-19 household transmission team, comparison of home antigen testing with RT-PCR and viral culture during the course of SARS-CoV-2 infection. JAMA Intern. Med. 2022 doi: 10.1001/jamainternmed.2022.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merckx J., Wali R., Schiller I., Caya C., Gore G.C., Chartrand C., Dendukuri N., Papenburg J. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann. Intern. Med. 2017;167:394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 38.Gökharman F.D., Ertem G.T., Aydın S., Büyükdemirci A., Yüksekkaya E., Tokur O., Koşar P.N. Evaluation of thorax computed tomographic findings in COVID-19 variant cases. Respir. Investig. 2022;S2212–5345(21):00215-X–X. doi: 10.1016/j.resinv.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available on request from the corresponding author.