Abstract
Emerging evidence has identified viral circular RNAs (circRNAs) in human cells infected by viruses, interfering with the immune system and inducing diseases including human cancer. However, the biogenesis and regulatory mechanisms of virus-encoded circRNAs in host cells remain unknown. In this study, we used the circRNA detection tool CIRI2 to systematically determine the virus-encoded circRNAs in virus-infected cancer cell lines and cancer patients, by analysing RNA-Seq datasets derived from RNase R-treated samples. Based on the thousands of viral circRNAs we identified, the biological characteristics and potential roles of viral circRNAs in regulating host cell function were determined. In addition, we developed a Viral-circRNA Database (http://www.hywanglab.cn/vcRNAdb/), which is open to all users to search, browse and download information on circRNAs encoded by viruses upon infection.
Keywords: viral circRNA, RNA-Seq, RNase R, host cells, Viral-circRNA Database
Data Summary
Public virus-infected RNA-Seq data were collected from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and summarized in Tables S1–S3 (available in the online version of this article). Raw sequencing data generated from our own laboratory were deposited under NCBI BioProject Accession PRJNA756819 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA756819/). We confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
Viral circular RNAs (circRNAs) have been recently identified in infected host cells, but recognizing virus-encoded circRNAs and understanding their functions in diseases is at an early stage. In this study, we systematically identified thousands of the virus-encoded circRNAs in virus-infected cancer cell lines and patients. Furthermore, the biological characteristics and potential roles of viral circRNAs in regulating host cell function were determined. Finally, virus-encoded circRNAs were deposited in the Viral-circRNA Database. Overall, the findings will provide novel insight into the world of viral physiology and pathology.
Introduction
Cancer is a genetic disease in which tumour cells grow uncontrollably and invade nearby tissues or spread to other parts of the body [1]. According to the Global Cancer Statistics 2020 [2], cancer was the leading cause of disease mortality, with 10 million deaths in 2020. Female breast cancer has been the most commonly diagnosed cancer with an estimated 0.69 million deaths (6.9 %), followed by lung, colorectal, prostate and stomach cancers. Viruses cause a variety of human diseases including malignancies [3]. It is estimated that Epstein-Barr virus (EBV) infects 90 % of the world’s population and is related to nasopharyngeal carcinoma (NPC), gastric cancer (GC), Burkitt lymphoma (BL), post-transplant lymphoproliferative disorder (PTLD) and natural killer (NK)/T cell lymphoma [4, 5]. Kaposi’s sarcoma-associated herpesvirus (KSHV) is a causative agent of primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD) [6]. Rhesus macaque lymphocryptovirus (rLCV), EBV and KSHV all belong to the gammaherpesvirus family and play a role in the occurrence and development of tumours [7]. Human papillomavirus (HPV), one of the most common viruses infecting the reproductive system, with more than 200 subtypes identified, is responsible for most cervical cancers and leads to vulvar cancer, penile cancer, vaginal cancer, anal cancer and oropharyngeal cancer [8–10].
Circular RNAs (circRNAs) are produced from pre-mRNA through back-splicing to form a covalently closed loop structure without a 5′-end cap and 3′-end poly(A) tail [11–13]. Viroids, single-stranded, covalently closed circular RNA molecules, were observed under an electron microscope in 1976 [14] and were the first ‘circular-form RNA’ discovered in nature. Of note, viroids are circular RNAs which are self-replicating, while circRNAs are classically understood as circular RNA transcripts expressed from genes. Since then, some studies have identified several circular transcripts and proposed their underlying mechanisms [11, 15]. Since about 2010, when high-throughput sequencing was burgeoning, thousands of circRNAs have been detected as being expressed in eukaryotes [16, 17], functionally modulating molecular activities by sponging micro RNAs (miRNAs) [18], interacting with RNA binding proteins (RBPs) [12], affecting parental gene expression [19] and encoding polypeptides [20]. Moreover, circRNAs may serve as promising biomarkers for human diseases due to their stability, conservation and high abundance in body fluids [21].
In recent years, virus-encoded circRNAs have been reported to be expressed and functional in virus-infected human tissues or cell lines [22–28]. These studies presented new research opportunities for understanding viral non-coding RNA and viral oncogenesis. EBV circRNAs may act as human miRNA sponges during viral infection, the cell cycle and oncogenesis [29]. HPV E7 protein can be translated by N6-methyladenosine (m6A)-modified circE7 to promote cancer cell growth [27], as m6A recruits the ribosome to mediate translation initiation from the middle of the genome [30]. However, it remains a major challenge to recognize virus-encoded circRNAs and understand their functions in host cells.
According to two independent benchmarking studies on circRNA detection tools [31, 32], CIRI [33], CIRCexplorer [34] and KNIFE [35] achieved excellent performance in balancing sensitivity and precision. Moreover, because viral genomes are not well annotated, de novo circRNA detection tools (such as CIRI2 [36], find_circ [37] and circRNA_finder [38]) that do not depend on genome annotation were considered. In this study we used CIRI2 [36] to systematically identify circRNAs of viral origin based on RNase R (a 3′exonuclease that digests linear RNAs)-treated and circRNA-enriched RNA-Seq data from cancer samples.Based on the viral circRNAs identified by CIRI2, we analysed and annotated these circRNAs to obtain systematic insight into the characteristics of viral circRNAs. We further developed a database for viral circRNAs to facilitate the retrieval of information related to viral circRNAs to explore their roles in host cells.
Methods
Analysis workflow
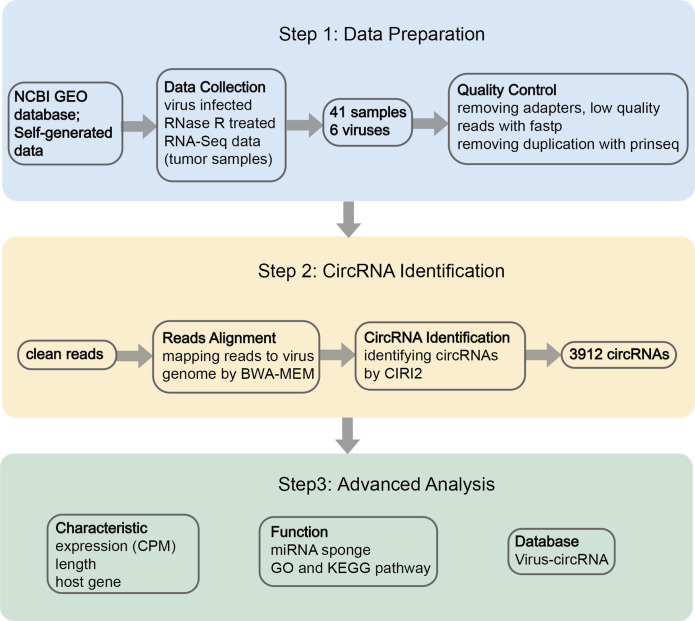
The analysis workflow is outlined in Fig. 1. First, RNase R-treated RNA-Seq samples were collected, and raw reads were preprocessed by a quality control pipeline (Step 1). Then, the circRNA detection tool CIRI2 was used to identify viral circRNAs (Step 2). Finally, the viral circRNAs were annotated, analysed and stored in the Viral-circRNA database.
Fig. 1.
An analysis pipeline to identify viral circRNAs from RNase R-treated RNA-Seq data of virus-infected tumour samples. The schematic depiction shows the three steps of the workflow, which include data collection and processing, identification of viral circRNAs, and data analysis and construction of the Viral-circRNA Database.
RNA-Seq
Total RNA-Seq or RNase R-treated RNA-Seq data are suitable for circRNA identification. To control for false positives, only RNase R-treated RNA-Seq samples were taken to identify viral circRNAs. Virus-infected and RNase R-treated RNA-Seq data were downloaded from the NCBI GEO [39] and covered six viruses, EBV, KSHV, rLCV, Middle East respiratory syndrome coronavirus (MERS-CoV), HPV and hepatitis B virus (HBV) (Table S1). Six poly(A)-selected tumour datasets infected by viruses included in our study (Table S2) were downloaded from the GEO database for false discovery rate (FDR) estimation.
Additionally, three RNase R-treated RNA-Seq samples, including two HPV-infected samples from patients with cervical cancer and one HBV-infected liver cancer cell line, Hep3B, were generated by our own laboratory. Human cervical cancer samples were collected from Tongji University Shanghai East Hospital. The raw and processed sequencing data were submitted to the SRA database (https://www.ncbi.nlm.nih.gov/sra) [40] under accession number PRJNA756819.
RNA quantification, sample preparation, library construction and Illumina sequencing
RNA degradation and contamination were monitored on 1 % agarose gels. RNA purity was checked using a NanoPhotometer spectrophotometer (IMPLEN). RNA concentration was measured using a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies), and RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies). For circRNA sequencing, ribosome-depleted and RNase R-treated RNA samples were used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s recommendations. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq platform, and 125 bp/150 bp paired-end reads were generated.
Ninety-six severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)-infected total or rRNA-depleted RNA-Seq samples across nine datasets (Table S3) were downloaded from the GEO database for additional analysis.
Processing of raw RNA-Seq data
The quality of sequencing reads was evaluated with FastQC v0.11.6 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [41]. Raw reads from the RNA-Seq data were processed by removing adapters and low-quality bases using FASTQ v0.21.0 [42] with default parameters and by reducing sequence duplication using PRINSEQ-lite v0.20.4 [43] with derep=1.
Identification of viral circRNAs
The clean reads were then aligned to a reference genome of the virus with BWA-MEM using parameter -T 19 recommended by the CIRI2 manual. CIRI2 v2.0.6 [36] was applied with default parameters to identify viral circRNAs. The viral circRNA with at least two junction reads in its back-splicing site was initially identified. After basic statistics, to control for false positives, circRNAs with at least five junction reads were selected for further analysis. CircRNAs with the same back-splicing site identified from different samples were integrated as one circRNA.
Characterization of viral circRNAs
In the next step, these viral circRNAs were analysed in terms of their host genes, their length, transcription strand, the number of junction reads and their expression levels. The expression level of a viral circRNA was measured by the natural logarithm of counts per million mapped reads (CPM), which was calculated as follows: , where n and N represent the number of junction reads and the number of mapped reads, respectively.
Function analysis of viral circRNAs
CircRNAs with junction reads ≥5 and with lengths ≤1 kb and human miRNAs from miRBase [44] were collected to predict miRNA–circRNA interactions using miRanda v3.3a [45] and TarPmiR [46]. To control for false positives, strict parameter settings were used for the prediction, and only interactions identified by both tools were accepted for further analysis. The parameters for miRanda were Max Score ≥160 and Max Energy≤−20 kcal mol–1, and for TarPmiR, they were Binding Probability=1 and Energy≤−20 kcal mol–1.
To measure the quality of the predicted miRNA–circRNA interactions, the outputs of miRanda including two metrics – Max Score and Max Energy – as well as the output of TarPmiR – Energy – were integrated to calculate a combined quality score. Max Score, Max Energy and Energy were respectively scaled to between 0 and 1 by min-max normalization. Because lower energy means a more thermodynamically stable binding, Max Energy in miRanda and Energy in TarPmiR were inverted before scaling. The combined quality score was defined as the average of the scaled outputs of miRanda and TarPmiR, with higher scores indicating higher quality. Based on miRNA–circRNA interactions, the Cytoscape 3.9.1 software [47] was used to build an interaction network in which the width of edges was determined by the combined quality score.
Furthermore, miRNA–mRNA interactions were retrieved from the miRNA target databases TargetScan [48], miRDB [49] and miRTarBase [50]. Finally, Gene Ontology (GO) [51] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [52] pathway enrichment analyses were conducted on the overlapping miRNA targets shared by the three databases using the R package clusterProfiler v3.18.1 [53]. All the KEGG pathways and GO terms with q-values <0.05 were considered as significant enrichment.
Graphical visualization
The R package circlize v0.4.12 [54] were used to visualize viral circRNAs. Viral circRNAs with abundant isoforms were plotted by SpliceV [55]. The coverage curve of circRNAs on the viral reference genomes were generated by Gviz v1.34.1 [56].
Development of the Viral-circRNA Database
The Viral-circRNA Database (http://www.hywanglab.cn/vcRNAdb/) was built on the Django (version 2.2.7) framework and run on the Apache 2 web server with MySQL (version 5.7.24) as the database engine. The JavaScript libraries Echarts and Highcharts were used for drawing graphs. The database is available online without registration and was optimized for most prevalent browsers, such as Chrome.
Statistical analysis
The Benjamini and Hochberg procedure was used to calculate adjusted P-values in GO and KEGG enrichment analyses. The Wilcoxon rank-sum test was used to compare the ln(CPM) values between long and non-long circRNAs. All statistical analyses were executed in R.
Results
Data collection and identification of viral circRNAs
To identify circRNAs of viral origin in host cells, we interrogated RNase R-treated RNA-Seq samples from virus-infected cell lines and patients with lymphoma, lung adenocarcinoma, cervical carcinoma and gastric carcinoma (Figs. 1 and 2a) from the NCBI GEO/SRA database. Among them, 69 % (27 of 39) of samples were from lymphoma patients, including the following subtypes: rhesus macaque lymphoma, primary effusion lymphoma, Burkitt’s lymphoma and B cell lymphoma. In addition, three supplementary RNase R-treated RNA-Seq samples were generated by our own laboratory, including two HPV-infected samples from patients with cervical cancer and one HBV-infected liver cancer cell line, Hep3B. In total, our analysis included 41 cancer samples (Table S1) infected with six types of viruses: KSHV, EBV, rLCV, MERS-CoV, HBV and HPV.
Fig. 2.
Statistical data of viral circRNAs. (a) Pie chart showing the distribution of cancers included in this analysis. The subgraph of the pie chart shows the subtypes of lymphoma. (b) The number of viral circRNAs identified from five viruses, MERS-CoV, KSHV, EBV, rLCV and HPV18. (c) The distribution of the number of junction reads for detecting viral circRNAs from each sample. (d) The distribution of circRNA expression measured by the natural logarithm of CPM. (e) The distribution of viral circRNA length. (f) The number of circRNAs identified across the samples. Graphical representation of EBV (g), KSHV (h), MERS-CoV (i) and rLCV (j) circRNAs using the program circlize. The outer circle of the viral genome is arranged in clockwise order, with each arch connecting the starting position and ending position of a single circRNA in the inner circle. The gradient colour of the arches corresponds to the expression level of circRNAs, characterized by the natural logarithm of the average CPM for each circRNA. Both positive- (red) and negative- (blue) strand circRNAs are shown. The purple curve and blue curve in the middle circle indicate the number of viral circRNAs for which each nucleotide is covered on the plus strand and minus strand, respectively.
Raw reads from the RNA-Seq data were processed by removing low-quality reads and adaptor dimers and by controlling sequencing duplication. After clean reads were aligned to the viral genome, the circRNA detection tool CIRI2 was used to identify 3912 viral circRNAs (Fig. 1). Among them, 3198 were from MERS-CoV, followed by 590 from KSHV, 102 from EBV, 19 from rLCV and three from HPV18 (Fig. 2b). To estimate the FDR, we collected the poly(A)-selected RNA-Seq datasets with at least one dataset for each cancer type infected with the viruses in our study (Table S2) and identified the circRNAs with the tool CIRI2. CircRNAs detected in both poly(A)-selected and RNase R-treated samples were considered to be false positives. Therefore, the estimated FDR was 0.0043 (17 of 3912).
Lengths and expression levels of viral circRNAs
The number of junction reads of circRNAs in each sample varied from two to 34 232, and 70 % (4139 of 5924) of the circRNAs had at least five junction reads (Fig. 2c), suggesting their reliability as true positives. Based on the number of junction reads, the expression levels of viral circRNAs were measured as CPM. Of note, the expression levels of 45 % (2694 of 5924) of the circRNAs were greater than 1, and 12 % (714 of 5924) reached up to 10, showing that a small portion of circRNAs had relatively high expression levels (Fig. 2d). For circRNAs with the same back-splicing site identified from different samples, we integrated them as one circRNA. The statistics also showed that 50 % (1963 of 3912) of circRNAs ranged from 200 to 500 bp in length, which was in line with the characteristics of circRNAs. Interestingly, 8 % (311 of 3912) of circRNAs had almost head-to-tail joining of viral genomes with lengths ≥10 kb (Fig. 2e). Among 3912 viral circRNAs, 2602 were only identified in one sample, 738 across two samples, 515 across three samples, 36 across four samples and 21 across at least five samples, suggesting that the circRNAs identified across more samples are more likely to be true positives (Fig. 2f). However, the circRNAs only identified in one sample were not necessarily false positives due to the limited sample size.
To further control for false positives, we selected circRNAs with at least five junction reads covering the back-splicing sites as high-confidence viral circRNAs for the following analysis. As a result, the estimated FDR of these circRNAs declined to 0.0023 (six of 2612). We created circular diagrams to display back-splicing sites (inner circle) and coverage of junction reads (middle circle) collectively on the viral genome (outer circle) (Fig. 2g–j). In the inner circle, the starting position and ending position of a circRNA are connected by a curve, with red and blue representing the positive strand and negative strand, and darker colours implying higher expression. EBV circRNAs were ubiquitously highly expressed (Fig. 2g). In the middle circle, circRNAs expressed from the positive strand and negative strand are shown in purple and blue, respectively, with the y-axis representing the coverage of junction reads. In EBV (Fig. 2g), KSHV (Fig. 2h) and rLCV (Fig. 2j), the circRNAs were primarily expressed on the positive strand. However, in MERS-CoV (Fig. 2i), more circRNAs were expressed on the negative strand. Moreover, in EBV, MERS-CoV and rLCV (Fig. 2g, i, j), interestingly, there were some arrow-pointing circRNAs with back-splicing sites almost joining across the entire genome in a head-to-tail manner, demonstrating the long-range cyclization of genomic RNAs.
Host genes of viral circRNAs
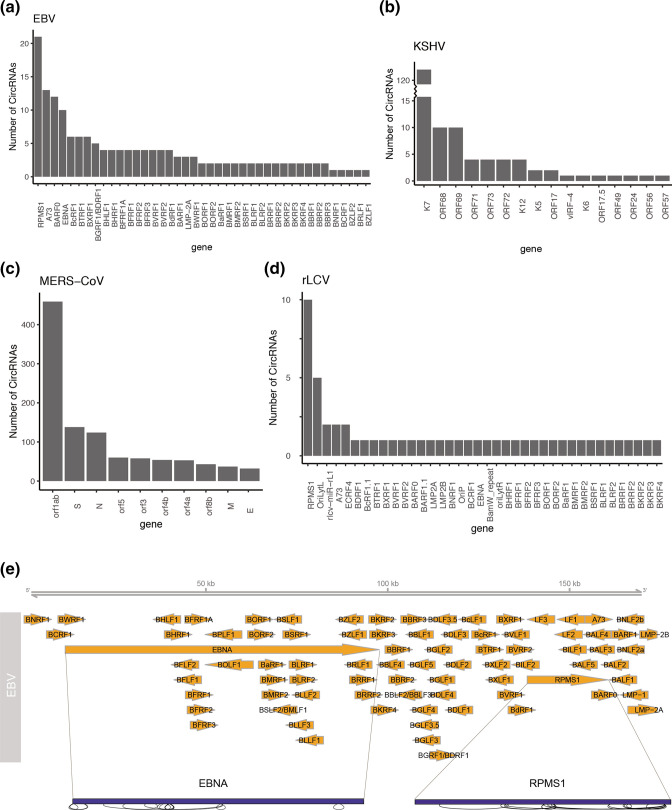
To observe the host genes from which viral circRNAs were transcribed, we annotated the circRNAs (junction reads ≥5) by connecting their genomic positions to loci of viral genes (Fig. 3). The results showed that one viral gene could produce multiple circRNA isoforms, with the viral genes RPMS1/A73/BARF0 and EBNA in EBV (Fig. 3a), K7 and ORF68/ORF69 in KSHV (Fig. 3b), orf1ab in MERS-CoV (Fig. 3c) and RPMS1 in rLCV (Fig. 3d) expressing the most circRNAs. For example, in EBV (Fig. 3a, e), 21 circRNA isoforms were transcribed from RPMS1, followed by 13 from A73, 12 from BARF0 and 10 from EBNA. However, because A73 and BARF0 have overlapping genome loci with RPMS1, some circRNAs are shared by these genes. In KSHV, 124 circRNA isoforms were transcribed from K7 (Figs 3b and S1a). Similarly, we observed an obvious peak at K7 (Fig. 2h), indicating abundant expression of K7 circRNAs in KSHV. Moreover, 459 circRNAs were transcribed from orf1ab, a gene covering approximately 60 % of the genome in MERS-CoV (Figs 3c and S1b). Two types of gammaherpesviruses, RPMS1 and rLCV, share a highly conserved transcript repertoire [57]. Remarkably, RPMS1 in EBV produced the most circRNA isoforms, and its orthologue in rLCV produced 10 different isoforms (Figs 3d and S1c). The circRNAs expressed from EBV RPMS1 have been reported to be relevant to tumour proliferation, apoptosis, invasion and metastasis [58]. Our combined results were in line with previous reports on viral circRNAs, such as circRNAs transcribed from RPMS1 [22, 26, 28], EBNA [26], BHRF1 [26], LMP2 [22, 26] and BHLF1 [26] in EBV; K7 [22, 25] and vIRF-4 [22, 23, 25] in KSHV; and RPMS1 and EBNA in rLCV [23]. These circRNAs are worthy of further investigation to explore their potential functions in cancers.
Fig. 3.
Viral genes originating from EBV (a), KSHV (b), MERS-CoV (c) and rLCV (d). The number of circRNAs that overlap with each viral gene is shown in the bar plot, which reflects the number of circRNA isoforms transcribed from each gene. (e) Graphical representation of EBV circRNAs with abundant isoforms made by SpliceV. Orange arrows demonstrate the genome structure of each virus. Genome regions outlined with black solid lines depict different circRNA isoforms generated from those regions.
Gammaherpesviruses such as EBV are involved in conversion of the infection state between latent and lytic phases. During much of its lifecycle, EBV remains silenced in the latent state to escape the host’s immune system, with only latent genes being expressed. During lytic infection, EBV switches to an active state, leading to extensive gene expression with the production of infectious viruses [59]. To evade immune recognition, EBV establishes different latent gene expression patterns consisting of EBNA, LMP, BART and an untranslated RNA called EBER [60]. As most of the EBV samples in our analysis were in a latent state [26], we observed that the major EBV circRNAs were transcribed from latent genes, such as EBNA, LMP and RPMS1/A73/BARF0 (Fig. 3a), indicating that these circRNAs might mediate immune escape similar to their linear transcripts. By comparison, most KSHV samples were in a lytic state, so KSHV circRNAs were primarily transcribed from lytic genes such as K7, ORF68 and ORF69, and only a small proportion of them were from latent genes, including ORF71, ORF72, ORF73 and K12 [61] (Fig. 3b). Significantly, K7, as the gene producing the most isoforms in KSHV, shares its locus with a long non-coding polyadenylated nuclear (PAN) RNA, which is the most abundant viral transcript during lytic replication [62]. Together, the expression of viral circRNAs is dependent on the infection state when host genes are activated. Therefore, as a supplement to linear transcripts, viral circRNAs may regulate the molecules of both viruses and infected cells because of their circular conformation and complementary sequence with linear RNAs to finally escape antiviral sensing of the host immune system in the latency state and manufacture more viruses in the lytic state.
Viral circRNAs as miRNA sponges
It is well known that endogenous circRNAs can function as miRNA sponges in eukaryotes [18]. To observe whether viral circRNAs can act as sponges to bind the miRNAs of the infected host cells, we used the miRNA target prediction tools miRanda (v3.3) [45] and TarPmiR [46] to predict the interactions between circRNAs and host miRNAs. We implemented this analysis on the circRNAs (length <1 kb and junction reads ≥5) and host miRNAs from the comprehensive miRNA database miRBase [44]. Based on the strict parameter settings (see Methods), our analysis identified 89 miRNA–circRNA pairs with 21 unique human miRNAs that were predicted by both tools.
To identify the important miRNAs with strong interaction to viral circRNAs, we integrated the outputs of miRanda including two metrics – Max Score and Max Energy – as well as the output of TarPmiR – Energy – to calculate a combined quality score of the interaction. Then we built a network in which wide edges represented a high quality score, and we focused on those hub miRNAs with multiple connections to circRNAs. The results showed that hsa-miR-6848–5p interacted with 21 KSHV circRNAs (Fig. S2a); hsa-miR-3085–5p (Fig. S2b), hsa-miR-6858–5p (Fig. S2c) and hsa-miR-8063 (Fig. S2d) respectively interacted with 18, 10 and nine MERS-CoV circRNAs; and hsa-miR-6747–5p interacted with four MERS-CoV circRNAs and one KSHV circRNA (Fig. S2e). More importantly, five MERS-CoV circRNAs interacting with hsa-miR-6747–5p had very high quality scores showing wide edges in the sub-network (Fig. S2e), providing more solid evidence for the potential role of hsa-miR-6747–5p in viral circRNA regulation. Other miRNAs with more than two interactions were shown in Fig. S2(f–k). According to the literature, hsa-miR-8063 was found to inhibit pancreatic cancer progression through inducing inhibition on ZFP91 [63], to suppress the epithelial–mesenchymal transition of lung cancer cells by regulating its target gene MAGAT3 [64], and to induce cell migration through inhibition of the tumour suppressor FOXN2 [65]. Hsa-mir-4745–5p was strongly dysregulated in Parkinson’s disease (PD) and can be directly regulated by HNF4a [66], which was identified as a longitudinal PD blood biomarker [67]. Hsa-mir-4745–5p was also observed to be upregulated in patients with colorectal cancer [68]. These all suggest there are potential interactions between host miRNAs and viral circRNAs, and some miRNAs deserve further investigation for their potential role in the regulation of viral pathology.
To further explore the functions of viral circRNAs, we used three miRNA target databases, TargetScan [48], miRDB [49] and miRTarBase [50], to retrieve the target genes of the 21 miRNAs above. This analysis led to the identification of 638 miRNA–mRNA interactions and 604 target genes. Enrichment of target genes based on KEGG pathway and GO analyses showed significantly enriched pathways and functions (Fig. 4). The top 10 enriched KEGG pathways showed a strong correlation with viruses and cancer (Fig. 4a), suggesting that viral circRNAs in tumour samples participate in viral infection and tumorigenesis. GO-enriched terms provided clues to how viral circRNAs play regulatory roles in the infected host cells. Viral circRNAs may regulate the cell cycle and protein structure and localization (Fig. 4b) through transcription repressor activity (Fig. 4c). In addition, cellular component terms, including nuclear envelope and nuclear speck, were enriched (Fig. 4d). Overall, the enriched functions were in line with the roles of viruses, indicating that some viral circRNAs function as host miRNA sponges to regulate infected host cells.
Fig. 4.
Functional analysis and negative strand bias of viral circRNAs. KEGG pathway and GO enrichment analyses of viral circRNA interacting genes. Only the top 10 enriched terms of the KEGG pathway (a), biological process (BP) (b), molecular function (MF) (c) and cellular component (CC) (d) are shown. The coverage curve of circRNAs on the reference genomes of MERS-CoV (e) and SARS-CoV-2 (f). Genes with known mapped positions in the genome are shown in orange. The pink curve and blue curve show the number of viral circRNAs for which each nucleotide is covered on the plus strand and minus strand, respectively.
Negative strand bias of viral circRNAs in coronavirus
There were 3198 viral circRNAs identified in MERS-CoV, accounting for 82 % of the total in this study (Fig. 2b). These circRNAs were identified from one study in which the human lung cancer cell line Calu-3 was infected with MERS-CoV for 6 and 24 h in vitro, followed by RNase R-treated RNA-Seq. Our analysis identified dozens of circRNAs in cell lines at 6 h post-infection and thousands of circRNAs at 24 h post-infection. This result suggested that fast replication of the virus in the host cells was accompanied by the production of abundant viral circRNAs. Therefore, we considered that this cell line provided a good system to study viral circRNAs post-infection.
To gain more insight into the characteristics of viral circRNAs, we extracted viral circRNAs from MERS-CoV and observed the distributions of junction reads, expression levels and lengths (Fig. S3a–c), which were similar to those identified from the pooled viruses (Fig. 2c–e). Then, we focused on MERS-CoV circRNAs (junction reads ≥5) as strict false-positive controls. Interestingly, when mapping the junction reads of viral circRNAs onto positive-sense (+) and negative-sense (−) strands of the MERS-CoV genome (Fig. 4e), we found that circRNAs were widely expressed across the whole genome and were highly expressed in some regions of orf1ab, S, orf3/orf4a/orf4b and N/orf8b. Surprisingly, we observed that there were twice as many circRNAs on the negative-sense (−) strand as on the positive-sense (+) strand (Fig. 4e). As a positive-sense ssRNA virus, MERS-CoV utilizes full-length replication or discontinuous transcription to generate negative-sense RNA genomes or subgenomic negative-sense RNAs, which serve as a template for the synthesis of positive-strand RNAs rather than encoding proteins [69]. It is unclear whether these negative-sense circRNAs play regulatory roles in the process of positive-sense RNA synthesis.
Since these viral circRNAs were from in vitro experiments, technical factors and biological factors may also impact the process of RNA cyclization. To observe whether this negative-strand bias truly exists in positive-sense ssRNA viruses, we collected 96 SARS-CoV-2-infected total or rRNA-depleted RNA-Seq samples across nine datasets, instead of collecting the few RNase R-treated samples (Table S3), and identified 376 circRNAs. To control for false positives caused by RNA-Seq data without RNase R treatment, we narrowed down the circRNAs to those with junction reads ≥5, leading to 140 circRNAs for further analysis. The results showed that a negative-strand bias was still observed for viral circRNAs identified in SARS-CoV-2 (Fig. 4f). Together, these findings in MERS-CoV and SARS-CoV-2 reveal that positive-sense ssRNA viruses produce more abundant circRNAs encoded from the negative strand.
Long-range cyclization of the viral genome and subgenome
Although the lengths of circRNAs are hundreds of bases in most cases, in our analysis, after selecting circRNAs with at least five junction reads, there were still 9 % (191 of 2199) of the circRNAs in MERS-CoV and 5 % (two of 43) of those in EBV that were longer than half of the genome. This result suggested an interaction between 5′-terminal sequences and 3′-terminal sequences, causing a crosslink between two ends of the genome or subgenome, which leads to long-range cyclization.
The cyclization positions of all circRNAs in EBV and MERS-CoV are shown in Fig. 5(a, b). With the short-length grey circRNAs near the diagonal as the background, orange and red circRNAs located at the upper left corner were from long-range cyclization, covering ≥50 % and ≥90 % of the genome, respectively. We compared the expression levels between RNAs from long-range cyclization and short-length circRNAs in EBV (Wilcoxon rank-sum test P=0.018, Fig. 5c) and in MERS-CoV (Wilcoxon rank-sum test P<2×10−16, Fig. 5d) and observed a significantly higher expression of long circRNAs in MERS-CoV (Fig. 5d). Long-range cyclization was also observed in the SARS-CoV-2 samples described above (Fig. 5e and Wilcoxon rank-sum test P=0.049, Fig. 5f).
Fig. 5.
Long-range cyclization of the viral genome and subgenome. The scatter plot shows the cyclization positions of circRNAs in EBV (a) and MERS-CoV (b). The circRNA, formed by long-range cyclization, is highlighted in red when its length is ≥90 % of the reference genome and in orange when its length is ≥50 % but <90 %. Violin plot of ln(CPM) for EBV (Wilcoxon rank-sum test P=0.018) (c) and MERS-CoV (Wilcoxon rank-sum test P<2×10−16) (d) circRNAs grouped into long (length ≥50 % of the reference genome) and non-long (length <50 % of the reference genome) groups. (e) Scatter plot of the cyclization positions of SARS-CoV-2 circRNAs. The circRNA, formed by long-range cyclization, is highlighted in red when its length is ≥90 % of the reference genome and in orange when its length is ≥50 % but <90 %. (f) Violin plot of ln(CPM) for SARS-CoV-2 circRNAs grouped into long (length ≥50 % of the reference genome) and non-long (length <50 % of the reference genome) groups (Wilcoxon rank-sum test P=0.049).
A database for viral circRNA in host cells
To enable knowledge sharing and reuse, we developed the Viral-circRNA Database (http://www.hywanglab.cn/vcRNAdb/, Fig. S4), which provides comprehensive knowledge of viral circRNAs identified in cancer samples to support further exploration of their physiological and pathological roles in host cells. The web interface of Viral-circRNA is user-friendly and allows users to search (Fig. S4a), browse (Fig. S4b) and download data. The database includes the following information on viral circRNAs: virus origin, reference genome, starting position, ending position, length, expression level, host gene, cancer type, RNA-Seq sample and visualization of the back-splicing position (Fig. S4c). In addition, an overall view of circRNAs produced from a given virus (Fig. S4a) and a data summary and basic statistics of the database are available (Fig. S4d). Links to external databases are also provided.
We performed a side-by-side comparison with VirusCircBase [70], a published viral circRNA database, from the perspective of sample type, RNA-Seq library, detection tool, etc. (Table S4). There were 3247 overlapped circRNAs which occupy 83 % (3247 of 3912) and 7 % (3247 of 46440) of the circRNA collection in our database and VirusCircBase database, respectively. Our analysis only focuses on tumour samples and carefully controls the false positives, by just taking advantage of RNase R-treated samples and employing the circRNA detection tool CIRI2, which has been shown to achieve excellent performance and is appropriate for viral genome detection. Therefore, our database provides less but more reliable circRNAs for promoting research in virus-infected tumours. More importantly, we provide several fields, including junction reads, supported samples, CPM, average junction reads across samples, etc. Users can filter the circRNAs based on their own criteria and download the data.
Discussion
In this study, we have used publicly available RNase R-treated RNA-Seq data of cancer cell lines and patients from public databases and self-generated data to systematically identify viral circRNAs in the context of cancers. Our study identified 3912 viral circRNAs encoded by viruses. Further analysis revealed that 50 % of the viral circRNAs ranged from 200 to 500 bp in length, and the circRNAs were intensively transcribed from viral genes such as RPMS1 in EBV and K7 in KSHV. Interestingly, we identified some circRNAs with lengths longer than half of the genome, which provides supporting evidence of long-range cyclization of the viral genome and subgenome. Moreover, approximately 70 % of viral circRNAs were from negative strands of MERS-CoV (1537 of 2199) and SARS-CoV-2 (101 of 140). In addition, we built a viral-circRNA database to support researchers in further exploring the functions of viral circRNAs. Recent studies have shown that viruses such as EBV [22, 26], KSHV [24, 25], rLCV [23], HPV [27] and HBV [28] encode circRNAs. Increasing evidence suggests that some of them are functional in the infected host cells, opening a new field of research on non-coding RNAs. For example, EBV circRNAs may act as human miRNA sponges [29], and HPV circE7 could translate oncoprotein E7 [27]. Through systematic identification and characterization of viral circRNAs in host cells, our work has added knowledge of the presence of viral circRNAs, delved into their intriguing characteristics and potential functions, and explored their pathogenic role in virus-caused diseases.
Recent studies into circRNA biogenesis have mainly focused on eukaryotes [71]. Although the functions of most circRNAs in eukaryotes are not well explored, the known functions include molecular sponges of miRNAs [18], binding proteins to form circribonucleoproteins (circRNPs) [12], regulation of transcription and splicing [19], and regulation of the translation of polypeptides [20]. With the increasing evidence of viral circRNAs in human diseases [22, 25, 26], their functions need to be explored to improve our understanding of viral pathology. It has been reported that viral circRNAs suppress nasopharyngeal carcinoma cell proliferation and metastasis as miRNA sponges, similar to their functions in eukaryotes [58, 70]. To gain insight into such functions, first, we predicted target miRNAs of the viral circRNAs and observed their functions by enrichment analysis. The enriched functions were associated with cancer development, such as that of breast cancer, gastric cancer and lung cancer (Fig. 4a). Cellular component terms included several nuclear components (Fig. 4d). On the one hand, some of the circRNAs in eukaryotes accumulate in the nucleus and regulate transcription [19, 72]. On the other hand, it has been reported that the eukaryotic long non-coding RNA (lncRNA) MALAT1 localizes in nuclear specks [73] and lncRNA NEAT1 is essential for organization and integrity of nuclear paraspecks [74]. Both lncRNAs and circRNAs can act as miRNA sponges [18, 75], so circRNAs may indirectly work in nuclear events by competing with lncRNAs. Overall, our results indicated that some viral circRNAs were involved in carcinogenesis and may by acting as miRNA sponges and participating in lncRNA/circRNA–miRNA–mRNA regulatory networks, similar to eukaryote circRNAs [18]. Second, we investigated the host viral genes that contained the transcribed elements of circRNAs. Of note, the host genes for the highly expressed circRNAs or those with multiple circRNA isoforms, such as EBV RPMS1, EBNA and LMP2, mostly function in viral infections and evading immune responses (Fig. 3), suggesting that their encoded circRNAs may be involved in the viral life cycle. Moreover, as non-coding RNAs, viral circRNAs are non-immunogenic to the adaptive immune system. Recent research has shown that artificially designed GFP-tagged circRNAs are not recognized by innate immune receptors and do not trigger the type I IFN response [76], and that endogenous circRNAs tend to form 16–26 bp, imperfect RNA duplexes and act as inhibitors of dsRNA-activated protein kinase (PKR) related to innate immunity [77]. However, this field is still in its early stage. Further investigations are needed to reveal whether the antiviral sensing machine is effective for viral circRNAs.
Additionally, our results showed that in the coronaviruses MERS-CoV and SARS-CoV-2, there were more circRNAs encoded from the negative strand than from the positive stand. The negative-strand bias also existed in miRNA–circRNA interaction pairs identified by two miRNA target prediction tools, where 91 % (50 of 55) of MERS-CoV circRNAs predicted to interact with miRNAs were on the negative strand. It is well known that for positive-sense ssRNA viruses such as MERS-CoV, the negative-sense RNA genomes or subgenomic RNAs serve as templates for the synthesis of positive-strand RNAs [78]. We hypothesize that these negative-sense circRNAs may have regulatory roles in the process of positive-sense RNA synthesis and may function as miRNA sponges. However, the mechanism of negative-sense circRNAs remains unclear, and future studies are warranted to reveal their roles in viral biology and tumorigenesis.
Most circRNAs in humans are hundreds of bases in length. Our analysis showed that some of the identified circRNAs (9 % in MERS-CoV, 5 % in EBV) could be generated from both the entire genome and the subgenome, in a long-range form (Fig. 5a, b), which indicated long-range cyclization of the viral genome. Interestingly, the analysis based on SARS-CoV-2-infected total or rRNA-depleted RNA-Seq samples produced more convincing evidence of long-range cyclization (Fig. 5e). Moreover, the expression levels of RNAs with long-range cyclization were higher than those of short-length circRNAs (Fig. 5d, f). It has been reported that in HCV [79], flavivirus [80] and poliovirus [81], a long-range RNA–RNA interaction leads to genome cyclization, which is required for virus replication. In addition, in coronaviruses, 5′−3′-end crosstalk is a strategy for negative-strand subgenome RNA synthesis [82]. However, the genome cyclization process mentioned above refers to base pairing between 5′−3′-end complementary sequences rather than the long circRNAs we detected in this study. These viral circRNAs are independent circles that may function as regulatory elements through transcription-regulating sequences (TRS-B), which precede each viral gene [69]. Moreover, all of the coronaviruses transcribed subgenomic RNAs that started from the 3′ end but had different ending genes on the 5′ end; however, only the 5′-most ORF is translated in a cap-dependent manner [83]. Therefore, coronavirus transcripts are relatively long and tend to produce long circRNAs. Note that the synthesis of RNA by coronaviruses is totally different from that by eukaryotes, and the biogenesis of their circRNAs cannot be simply attributed to alternative splicing. The specific roles of coronavirus circRNAs are worthy of further study.
In this study, we performed rRNA depletion and RNase R-treated RNA-Seq on two HPV-infected patients with human cervical cancer and one HBV-infected human liver cancer cell line, Hep3B. Linear RNAs were degraded by exonucleases, and only circRNAs were left for sequencing. For the patients with cervical cancer, although no junction reads containing the back-splicing sites of circRNAs were found, reads that mapped to the viral genome still supported the presence of viral circRNAs. For instance, for the first patient, 98 % (12750 of 13007) of the reads that mapped to HPV genome subtypes were from the HPV71 genome (Fig. S5a). For the second patient 48 % (24588 of 50976), 20 % (9976 of 50976) and 15 % (7603 of 50976) of the mapped reads were mapped to HPV71, HPV18 and HPV16, respectively (Fig. S5b), showing a mixed infection of multiple HPV subtypes. However, no HBV-encoded circRNAs were detected in the HBV-infected cell line Hep3B. Together, this indicates that viral circRNAs were produced in HPV-infected patients with cervical cancer.
Despite the encouraging results, our study suffers from the following limitations, which we hope to address in future studies. First, our analysis is based on dozens of RNase R-treated and virus-infected samples. With the availability of more and better quality data, some findings will be more convincing. Second, the assumptions on the functions and production mechanisms of viral circRNAs were driven by data, and experimental validations should be confirmed to further understand their properties and mechanisms in host cells. Third, the number of circRNAs across viruses varies greatly. On the one hand, it may be affected by the biological factors, such as circRNA biogenesis and potential regulatory roles in viruses. For example, in EBV, the transcripts are produced through alternative splicing as in eukaryotes, while in coronavirus, the transcripts are generated in a discontinuous way to form subgenomic RNA. The different biogenesis of circRNAs may thus lead to the difference in number of circRNAs across the viruses. On the other hand, technical factors such as sample size and experimental conditions including reagent treatment, tissue or cell line, virus multiplicity of infection and infection time may also lead to the varied number of circRNAs across the viruses. For example, more circRNAs were identified in in vitro samples long times after virus infections. To sum up, both technical factors and biological factors can impact the process of RNA cyclization. With increasing diversity of data for technical and biological factors available, we will have a more comprehensive landscape on virus-encoded circRNAs in host cells.
In summary, our study utilized high-throughput RNase R-treated RNA sequencing data of virus-infected cancer samples to systematically identify viral circRNAs. Furthermore, our analyses based on these circRNAs provide a comprehensive view of the characteristics of circRNAs, including their expression levels, the lengths and the host genes, as well as their potential roles in regulating host cell function. In particular, we report some interesting findings, such as long-range cyclization of viral genomes and subgenome, and negative strand bias of viral circRNAs in coronavirus. Finally, we developed a Viral-circRNA Database, which is open to all users to search, browse and download information on circRNAs encoded by viruses upon infection, for promoting research into virus-infected tumours. Overall, the findings provide novel insight into the world of viral physiology and pathology. Our study sheds light on the prospects of viral circRNAs in virus biology and diseases. We believe that the study of viral circRNAs should focus more on the behaviour of circRNAs in viruses, which is different from that in eukaryotes. Therefore, studies integrating circRNA biology and virology will promote a better understanding of the group of viral circRNAs. Moreover, understanding the viral utilization of circRNAs will advance therapies in virus-infected diseases.
Supplementary Data
Funding information
This work was supported by grants from the National Natural Science Foundation of China (31771469), and the National Key Research and Development Program (2017YFC0908500).
Acknowledgements
We are grateful to the supercomputer centre of Bioinformatics of Tongji University for the allocation of computer time.
Author contributions
H.W. and Z.Y., conceived the hypothesis. S.C. and J.Z., designed and performed the data analysis and the database development. J.C., collected human cervical cancer samples. B.Z., X.T., Y.C., T.W., Y.X. and T.M., collected and preprocessed the data. S.C., J.Z., Z.Y. and H.W., wrote and revised the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study has been approved by the ethics committee of Tongji University Shanghai East Hospital (Ethics code: 2021-002). All participants signed an informed consent and the investigators obtained written informed consent prior to entry into the study.
Footnotes
Abbreviations: circRNA, circular RNA; CPM, counts per million mapped reads; EBV, Epstein–Barr virus; FDR, false discovery rate; HBV, hepatitis B virus; HPV, human papillomavirus; KSHV, Kaposi’s sarcoma-associated herpesvirus; lncRNA, long non-coding RNA; MERS-CoV, Middle East respiratory syndrome coronavirus; miRNA, microRNA; rLCV, Rhesus macaque lymphocryptovirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary figures and four supplementary tables are available with the online version of this article.
References
- 1.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8:e180–e190. doi: 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 4.Cohen JI. Epstein-Barr virus infection. N Engl J Med. 2000;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 5.Farrell PJ. Epstein-Barr virus and cancer. Annu Rev Pathol. 2019;14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023. [DOI] [PubMed] [Google Scholar]
- 6.Oksenhendler E, Boutboul D, Galicier L. Kaposi sarcoma-associated herpesvirus/human herpesvirus 8-associated lymphoproliferative disorders. Blood. 2019;133:1186–1190. doi: 10.1182/blood-2018-11-852442. [DOI] [PubMed] [Google Scholar]
- 7.Marshall VA, Labo N, Hao X-P, Holdridge B, Thompson M, et al. Gammaherpesvirus infection and malignant disease in rhesus macaques experimentally infected with SIV or SHIV. PLoS Pathog. 2018;14:e1007130. doi: 10.1371/journal.ppat.1007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araldi RP, Sant’Ana TA, Módolo DG, de Melo TC, Spadacci-Morena DD, et al. The human papillomavirus (HPV)-related cancer biology: An overview. Biomed Pharmacother. 2018;106:1537–1556. doi: 10.1016/j.biopha.2018.06.149. [DOI] [PubMed] [Google Scholar]
- 9.Taberna M, Mena M, Pavón MA, Alemany L, Gillison ML, et al. Human papillomavirus-related oropharyngeal cancer. Ann Oncol. 2017;28:2386–2398. doi: 10.1093/annonc/mdx304. [DOI] [PubMed] [Google Scholar]
- 10.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 11.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 12.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capel B, Swain A, Nicolis S, Hacker A, Walter M, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 16.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Huang C, Bao C, Chen L, Lin M, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 20.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Yang T, Xiao J. Circular rnas: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, et al. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci USA. 2018;115:E8737–e45. doi: 10.1073/pnas.1811728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ungerleider NA, Jain V, Wang Y, Maness NJ, Blair RV, et al. Comparative analysis of gammaherpesvirus circular RNA Repertoires: conserved and unique viral circular RNAs. J Virol. 2019;93 doi: 10.1128/JVI.01952-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, et al. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci USA. 2018;115:12805–12810. doi: 10.1073/pnas.1816183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abere B, Li J, Zhou H, Toptan T, Moore PS, et al. Kaposi’s sarcoma-associated herpesvirus-encoded circRNAs are expressed in infected tumor tissues and are incorporated into virions. mBio. 2020;11 doi: 10.1128/mBio.03027-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, et al. The Epstein Barr virus circRNAome. PLoS Pathog. 2018;14:e1007206. doi: 10.1371/journal.ppat.1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekiba K, Otsuka M, Ohno M, Kishikawa T, Yamagami M, et al. DHX9 regulates production of hepatitis B virus-derived circular RNA and viral protein levels. Oncotarget. 2018;9:20953–20964. doi: 10.18632/oncotarget.25104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao Y, Zhao X, Liu J, Yang W. Epstein-Barr virus circRNAome as host miRNA sponge regulates virus infection, cell cycle, and oncogenesis. Bioengineered. 2019;10:593–603. doi: 10.1080/21655979.2019.1679698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Fan X, Mao M, Song X, Wu P, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen TB, Venø MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13:e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 37.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 38.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database Collaboration The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–6. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S A FastQC A Quality Control Tool for High Throughput Sequence Data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 42.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enright AJ, John B, Gaul U, Tuschl T, Sander C, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding J, Li X, Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics. 2016;32:2768–2775. doi: 10.1093/bioinformatics/btw318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with cytoscape 3. Current Protocols in Bioinformatics. 2014;47:1–24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H-Y, Lin Y-C-D, Li J, Huang K-Y, Shrestha S, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 55.Ungerleider N, Flemington E. SpliceV: analysis and publication quality printing of linear and circular RNA splicing, expression and regulation. BMC Bioinformatics. 2019;20:231. doi: 10.1186/s12859-019-2865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahne DS, Ivanek R, Mueller A, Lianoglou S, Tan G. Gviz: plotting data and annotation information along genomic coordinates. R package version 1.6.0 ed2014
- 57.Rivailler P, Jiang H, Cho Y, Quink C, Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol. 2002;76:421–426. doi: 10.1128/jvi.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q, Shuai M, Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag Res. 2019;11:8023–8031. doi: 10.2147/CMAR.S218967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenzie J, El-Guindy A. Epstein-barr virus lytic cycle reactivation. Curr Top Microbiol Immunol. 2015;391:237–261. doi: 10.1007/978-3-319-22834-1_8. [DOI] [PubMed] [Google Scholar]
- 60.Kanda T. EBV-encoded latent genes. Adv Exp Med Biol. 2018;1045:377–394. doi: 10.1007/978-981-10-7230-7_17. [DOI] [PubMed] [Google Scholar]
- 61.Yan L, Majerciak V, Zheng ZM, Lan K. Towards better understanding of KSHV life cycle: from transcription and posttranscriptional regulations to pathogenesis. Virol Sin. 2019;34:135–161. doi: 10.1007/s12250-019-00114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conrad NK. New insights into the expression and functions of the Kaposi’s sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res. 2016;212:53–63. doi: 10.1016/j.virusres.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang Z, Ruan B, Zhong M, Xiong J, Jiang Y, et al. Silencing LINC00491 inhibits pancreatic cancer progression through MiR-188-5p-induced inhibition of ZFP91. J Cancer. 2022;13:1808–1819. doi: 10.7150/jca.65071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niu H, Qu A, Guan C. Suppression of MGAT3 expression and the epithelial-mesenchymal transition of lung cancer cells by miR-188-5p. Biomed J. 2021;44:678–685. doi: 10.1016/j.bj.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeong S, Kim SA, Ahn SG. HOXC6-Mediated miR-188-5p expression induces cell migration through the inhibition of the tumor suppressor FOXN2. Int J Mol Sci. 2021;23:9. doi: 10.3390/ijms23010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruf WP, Freischmidt A, Grozdanov V, Roth V, Brockmann SJ, et al. Protein binding partners of dysregulated miRNAs in Parkinson’s Disease Serum. Cells. 2021;10:791. doi: 10.3390/cells10040791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santiago JA, Potashkin JA. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson’s disease. Proc Natl Acad Sci U S A. 2015;112:2257–2262. doi: 10.1073/pnas.1423573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gungormez C, Gumushan Aktas H, Dilsiz N, Borazan E. Novel miRNAs as potential biomarkers in stage II colon cancer: microarray analysis. Mol Biol Rep. 2019;46:4175–4183. doi: 10.1007/s11033-019-04868-7. [DOI] [PubMed] [Google Scholar]
- 69.Sola I, Almazán F, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, et al. VirusCircBase: a database of virus circular RNAs. Brief Bioinform. 2021;22:2182–2190. doi: 10.1093/bib/bbaa052. [DOI] [PubMed] [Google Scholar]
- 71.Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 73.Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pisani G, Baron B. Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Noncoding RNA Res. 2019;4:128–134. doi: 10.1016/j.ncrna.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuang M, Zhao S, Jiang Z, Wang S, Sun P, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi: 10.1016/j.ebiom.2018.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67:228–238. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu C-X, Li X, Nan F, Jiang S, Gao X, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 78.Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 79.Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5’ and 3’ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/s1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lo CY, Tsai TL, Lin CN, Lin CH, Wu HY. Interaction of coronavirus nucleocapsid protein with the 5’- and 3’-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis. FEBS J. 2019;286:3222–3239. doi: 10.1111/febs.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.