Viruses and the discovery of Mediator
Just as biophysicists were identifying the component proteins of functional RNA polymerase II (RNAPII) complexes, virologists were identifying viral proteins that control it with peculiar efficiency [1]. Purified transcription components phosphorylate the RNAPII carboxyl-terminal domain (CTD) to initiate transcription, and this is enhanced by viral transactivators. In this way, viral proteins with their potent transcription activation domains (TADs) served as powerful tools for the characterization of DNA binding domains and TAD structures, e.g., the “acidic activator” (an acidic amphipathic alpha helix) and in the identification of host proteins that control transcription. The herpes simplex virus (HSV) transactivator, virion protein 16 (VP16, α-TIF, and Vmw65), was particularly valuable in transcription assays [1–3].
In the early 90s, prior to discovery of Mediator, a cyclin of unknown function, cyclin C, identified in screens for the rescue of yeast deficient in CLN genes (G1 cyclins) [4–6], was found to activate cyclin-dependent kinase 8 (CDK8) to phosphorylate the RNAPII CTD, placing it firmly at transcription control, not cell cycle [7–9]. At the same time, virologists discovered that VP16 and the adenovirus E1A proteins were associated with a large complex with cyclin C/CDK8 kinase activity [10].
This same period saw the sequence of an acutely transforming oncogenic retrovirus, walleye dermal sarcoma virus (WDSV) [11]. The WDSV genome encodes a retroviral cyclin (RV-cyclin) with only distant homology to any eukaryotic cyclin. The RV-cyclin rescued CLN-deficient yeast and induced hyperplastic lesions in mice carrying its transgene [12,13].
At this point, many components of Mediator had been identified as parts of a large, thyroid-hormone receptor complex (TRAP, THRAP, activator-recruited cofactor (ARC), and cofactor required for Sp1 activation (CRSP)) [14,15], and in 2002, the whole ARC/CRSP complex (Mediator) was purified by its VP16 affinity and its structure visualized by cryo-EM [16]. We showed that the RV-cyclin colocalized with transcription and splicing complexes in nuclei of mammalian cells and, in 2002, demonstrated its specific binding to and activation of human CDK8 (walleye and human CDK8 proteins are 98% identical) [17,18]. This was the first association of CDK8 with oncogenesis. In 2008, CDK8 was identified as an oncogene in a majority of human colon carcinomas and has since been implicated in a variety of human metastatic cancers [19,20].
The evolution of a viral cyclin that binds and activates the CDK component of Mediator illustrates the value of Mediator control for virus replication. While RV-cyclin is the only virus protein known to directly activate Mediator CDKs, the TADs of many viral transactivators function via direct contact with Mediator proteins, and additional virus protein contacts are being identified. Mediator offers refined control of host and virus gene expression, and viruses avail themselves of this control to meet their specific needs.
What is the Mediator complex?
RNAPII transcription is initiated by transcription factors bound to enhancer and promoter regions, recruitment of coactivator complexes that modify and remodel chromatin, and assembly of preinitiation complexes (PICs) with general transcription factors and RNAPII at core promoters. The Mediator complex, does just what it was named for: It mediates signals from transcription factors to assembled RNAPII complexes. Mediator is a bridge between a diverse array of DNA recognition factors and RNAPII. It also positions CDK7 at the RNAPII CTD for phosphorylation and transcription initiation [21–23]. Mediator coordinates and refines outputs from an array of disparate signals by reconfiguration of its components to control transcription initiation, elongation, and termination [24–26]. It is a cofactor in all RNAPII transcription [27,28].
Mediator is composed of approximately 30 proteins in 4 structural modules: the head, middle, and tail modules comprise the core, which can associate with the CDK8 kinase module (CKM) [29]. The Mediator core stabilizes the association of CDK7 in the general transcription factor TFIIH with the RNAPII CTD for phosphorylation of serine 5 in the CTD heptad repeats to initiate transcription [21–23,29–31]. The CKM is dissociable from Mediator. Its presence in the complex inhibits transcription initiation by blocking CDK7 access to the RNAPII CTD. However, the CKM is necessary for the release of paused polymerase complexes in response to specific enhancer-bound transcription factors that reprogram gene expression [29,32,33].
The CKM includes the CDK8-activating cyclin, cyclin C, and Mediator proteins, Med12 and Med13. MED12 increases CDK8 activation, and MED13 links the CKM with the core [31,34,35]. A CDK8 paralog, CDK19, present only in vertebrates, shares the kinase domain of CDK8 but diverges in the carboxyl-terminal tail. The CDK19 module includes cyclin C and Med12L and Med13L. CDK8 and CDK19 are highly conserved serine–threonine kinases that phosphorylate serines 2 and 5 in the heptad repeats of the RNAPII CTD to control transcription pausing and elongation, chromatin remodeling, and RNA processing and export [36–39]. CDK8 also phosphorylates itself, cyclin C, Med12 and Med13, Mediator middle and head modules, Cyclin H, multiple transcription factors, general transcription factors, the super elongation complex (SEC), chromatin remodelers, and serine 10 of histone H3 (H3S10) [8,9,40–43]. Autophosphorylation of the CKM yields full activation and controls CKM–Mediator association and Med13 degradation. Phosphorylation of Mediator middle and head modules controls the structure of core Mediator. Phosphorylation of Cyclin H, the CDK7 activator, inhibits transcription initiation. Phosphorylation of transcription factors controls reprogramming of gene expression, especially in response to metabolic, proliferative, and developmental signals by altering their activity or stability. Physical interactions between CKM–Mediator and pTEFb/SEC complexes at paused polymerases and SEC phosphorylation support pause release [44]. H3S10 phosphorylation by dissociated CKM promotes H3K9 demethylation and H3K14 acetylation and transcription activation [43].
CDK8 activity regulates nutrient stress response genes in yeast [45,46]. In metazoan organisms, CDK8 activity is necessary to reprogram cell identity and proliferation in response to stimuli like cytokines and serum and metabolism in response to cell stresses, such as hypoxia and starvation [36,37,42,47–50]. CDK8 controls transcription responses to Ras/Mitogen-activated protein (MAPK), Wingless/Integrated (WNT), transforming growth factor beta (TGFβ), nuclear factor kappa B (NF-κB), interferon gamma (IFNγ), and Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathways [42,51–53]. The CKM controls responses to a wide array of internal and external signals but has little control of steady-state house-keeping genes [32]. A possible selective mechanism for Mediator CKM dependent gene expression programs lies in the differential recruitment of elongation factors by the CKM versus Med26 of CKM-free Mediator complexes [29].
Ectopic RV-cyclin increases CDK8 kinase activity for histone H3 and RNAPII CTD in vitro. It also increases and extends the duration of CDK8 and RNAPII occupancy across the loci of serum response genes, prior to and after stimulation. This increases total mRNA levels from these CDK8-dependent genes and significantly increases cell proliferation [18,54–56].
Viruses target Mediator to activate viral gene expression
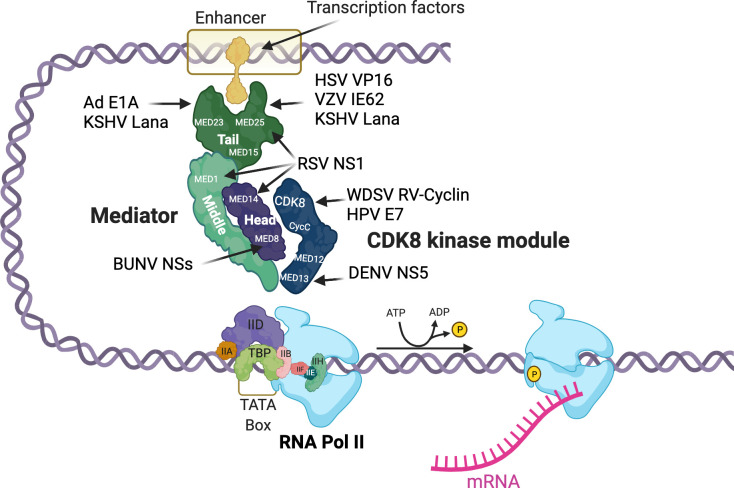
DNA viruses and retroviruses depend on host transcription machinery and encode proteins that control the extent and timing of their gene expression. This allows coordination of virus replication with capsid construction and packaging. The TADs of viral factors make high affinity contacts with Mediator proteins to activate and maintain high transcription rates of virus genes (Fig 1). These viral proteins served as models of host factors and led to the identification of specific Mediator protein structures, such as ACID, the activator interaction domain, in Med25, named for the VP16 acidic activator [57,58]. Other examples include the Kaposi’s sarcoma–associated herpesvirus Lana-1 protein, which binds Med15, Med23, and Med25 [59], the Varicella Zoster Virus IE62 protein, which targets Med25 [60], and the adenovirus E1A protein, which binds Med23 (SUR-2) [61]. For human papilloma virus 16 (HPV16), increased CDK8 occupancy on the long control region enhancer and late promoter in differentiating cells is associated with late promoter activity [62]. The WDSV RV-cyclin actually inhibits expression from the virus promoter during the latent phase of infection but carries a potent acidic TAD that activates host proliferative genes [63].
Fig 1. Viral proteins target the Mediator complex to control viral and host gene expression.
Viral proteins known to bind to or associate with Mediator proteins in the tail, middle, and head and CDK modules are depicted. Ad, adenovirus E1A; BUNV, Bunyamwera virus NSs; CDK8, cyclin-dependent kinase 8; DENV, dengue virus NS5; HPV, human papilloma virus E7; HSV, herpes simplex virus VP16; KSHV, Kaposi sarcoma–associated herpesvirus Lana-1; RSV, respiratory syncytial virus NS1; RV-cyclin, retroviral cyclin; VZV, varicella zoster virus IE62; WDSV, Walleye dermal sarcoma virus. Created with BioRender.com.
Viruses target Mediator to control cell differentiation and proliferation
Many viruses require host machinery to replicate their genome. For small DNA tumor viruses, polyoma, papilloma, and adenoviruses, host–cell proliferation provides DNA polymerase components that replicate viral DNA. Proteins encoded by these viruses target pRb and p53 to trigger cell cycle and block apoptosis and may also contact Mediator to control host gene expression, e.g., E1A [61].
The lymphotropic retroviruses and herpesviruses persist as integrated genomes or tethered episomes, so host cell proliferation replicates the viral genome. These viruses encode proteins that manipulate host transcription factors that contact Mediator tail proteins to promote cell division. Transition to virus production is also maintained via tight control of host transcription and requires Mediator.
A refined interplay between host proliferative response and virus latency and lytic replication is modeled in the seasonal development and regression of WDSV. Latent provirus produces low levels of RV-cyclin and OrfB transcripts. OrfB protein constitutively activates protein kinase C and AKT [64] to activate host immediate early genes, FOS, JUN, and EGR1. RV-cyclin increases CDK8 activity and chromatin occupancy of these genes to increase their rates of transcription elongation and reinititation [54]. After a season of tumor cell and provirus proliferation, the WDSV expression profile suddenly switches to genome replication, particle formation, and tumor regression timed with host spawning. Ectopic RV-cyclin alone can increase division even of highly proliferative human cancer cells [55]. This model ties CDK8 activity and chromatin occupancy to transcription elongation and reinitiation and to cell proliferation [54].
Viruses target Mediator for control of immune response
Viruses evolved many mechanisms to antagonize innate and adaptive immunity, especially interferon response pathways. Virus proteins that target transcription of key antiviral genes include those that contact Mediator. CDK8 and CDK19 regulate transcriptional responses to IFNγ [42,65], and CDK8 promotes release of paused RNAPII at IFNγ-induced genes [65]. Knockdown of cyclin C or CDK19 makes cells more sensitive to virus infection [42,65], indicating their elemental control of antiviral responses.
Orthobunyaviruses encode a nonstructural protein, NSs, which antagonizes the type I interferon response. Bunyamwera NSs binds Med8 in the head domain of Mediator to prevent phosphorylation of serine 2 in the RNAPII CTD and inhibit elongation of mRNA [66,67]. Human respiratory syncytial virus NS1, another antagonist of interferon signaling, contacts Med1, Med14, and Med25 and is enriched at enhancer regions, including enhancers of interferon-stimulated genes (ISGs), suggesting a role in their modulation [68]. HPV16 E7 also suppresses ISG transcription via CDK8, and knockdown of CDK8 in E6/E7-expressing cells increases ISG expression [69].
Viruses target Mediator for control of host cell metabolism
Virus infection frequently results in reprogramming of host cell metabolism in ways that support virus replication. As indicated above, Mediator, and CDK8 in particular, are key regulators of metabolic reprogramming. Dengue virus enhancement of glucose metabolism and oxidative phosphorylation is well characterized [70–72]. Infection induces CDK8 expression, and CDK8 activity is required for the up-regulation of metabolic genes [73]. The dengue virus RNA-dependent RNA polymerase, NS5, has high affinity for host chromatin and associates with Med13 [73,74] (personal communication, R. Perera). NS5 also interacts with spliceosomes to dysregulate splicing to favor virus replication and with polymerase associated factor 1 complex (PAF1C) to antagonize expression of PAF1-dependent immune response genes [75,76].
The question remains as to a specific role for NS5 and similar viral proteins in the direct control of host gene expression. There are many cell signaling pathways, triggered by virus infection and replication, which would result in metabolic reprogramming without benefit of direct virus control of Mediator. However, as we move into a new era of Mediator control of nuclear condensates to stabilize transcription and splicing components in response to cell signaling [52], we may reconsider the roles of viral proteins such as NS5 that have high concentrations on active chromatin [73,74]. Whether they guide or enhance reprogramming, viral proteins will once again serve as models for for investigations of eukaryotic gene expression.
Acknowledgments
Many laboratories have contributed to our knowledge of the Mediator complex and its roles during transcription. We appreciate these efforts and apologize to those whose work could not be cited due to space and reference limitations.
Funding Statement
This work was supported in part by NIH R01CA95056 (SLQ) and a Colorado State University College of Veterinary Medicine and Biomedical Sciences College Research Council Grant (SLQ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335(6190):563–4. doi: 10.1038/335563a0 [DOI] [PubMed] [Google Scholar]
- 2.Kristie TM, LeBowitz JH, Sharp PA. The octamer-binding proteins form multi-protein—DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989;8(13):4229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2(6):718–29. doi: 10.1101/gad.2.6.718 [DOI] [PubMed] [Google Scholar]
- 4.Lahue EE, Smith AV, Orr-Weaver TL. A novel cyclin gene from Drosophila complements CLN function in yeast. Genes Dev. 1991;5:2166–75. doi: 10.1101/gad.5.12a.2166 [DOI] [PubMed] [Google Scholar]
- 5.Leopold P, O’Farrell PH. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell. 1991;66:1207–16. doi: 10.1016/0092-8674(91)90043-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lew DJ, Dulic V, Reed SI. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–206. doi: 10.1016/0092-8674(91)90042-w [DOI] [PubMed] [Google Scholar]
- 7.Leclerc V, Leopold P. The cyclin C/Cdk8 kinase. Prog Cell Cycle Res. 1996;2:197–204. doi: 10.1007/978-1-4615-5873-6_19 [DOI] [PubMed] [Google Scholar]
- 8.Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene. 1996;12:2631–40. [PubMed] [Google Scholar]
- 9.Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18(4):1093–102. doi: 10.1038/sj.onc.1202399 [DOI] [PubMed] [Google Scholar]
- 10.Gold MO, Tassan JP, Nigg EA, Rice AP, Herrmann CH. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Res. 1996;24(19):3771–7. doi: 10.1093/nar/24.19.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holzschu DL, Martineau D, Fodor SK, Vogt VM, Bowser PR, Casey JW. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69(9):5320–31. doi: 10.1128/JVI.69.9.5320-5331.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lairmore MD, Stanley JR, Weber SA, Holzschu DL. Squamous epithelial proliferation induced by walleye dermal sarcoma retrovirus cyclin in transgenic mice. Proc Natl Acad Sci U S A. 2000;97(11):6114–9. doi: 10.1073/pnas.110024497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaPierre LA, Casey JW, Holzschu DL. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J Virol. 1998;72:8765–71. doi: 10.1128/JVI.72.11.8765-8771.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fondell JD, Guermah M, Malik S, Roeder RG. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci U S A. 1999;96(5):1959–64. doi: 10.1073/pnas.96.5.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12(3):127–34. doi: 10.1016/s1043-2760(00)00355-6 [DOI] [PubMed] [Google Scholar]
- 16.Taatjes DJ, Naar AM, Andel F III, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295(5557):1058–62. doi: 10.1126/science.1065249 [DOI] [PubMed] [Google Scholar]
- 17.Rovnak J, Casey JW, Quackenbush SL. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin). Virology. 2001;280:31–40. doi: 10.1006/viro.2000.0731 [DOI] [PubMed] [Google Scholar]
- 18.Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus cyclin interacts with components of the Mediator complex and the RNA polymerase II holoenzyme. J Virol. 2002;76:8031–9. doi: 10.1128/jvi.76.16.8031-8039.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455(7212):547–51. doi: 10.1038/nature07179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol. 2015;50(5):393–426. doi: 10.3109/10409238.2015.1064854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdella R, Talyzina A, Chen S, Inouye CJ, Tjian R, He Y. Structure of the human Mediator-bound transcription preinitiation complex. Science. 2021;372(6537):52–6. doi: 10.1126/science.abg3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rengachari S, Schilbach S, Aibara S, Dienemann C, Cramer P. Structure of the human Mediator-RNA polymerase II pre-initiation complex. Nature. 2021;594(7861):129–33. doi: 10.1038/s41586-021-03555-7 [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Yin X, Li J, Wu Z, Qi Y, Wang X, et al. Structures of the human Mediator and Mediator-bound preinitiation complex. Science. 2021;372(6546). doi: 10.1126/science.abg0635 [DOI] [PubMed] [Google Scholar]
- 24.Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in Mediator. Nat Struct Mol Biol. 2010;17(6):753–60. doi: 10.1038/nsmb.1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. Subunit Architecture and Functional Modular Rearrangements of the Transcriptional Mediator Complex. Cell. 2014;158(2):463. doi: 10.1016/j.cell.2014.06.036 [DOI] [PubMed] [Google Scholar]
- 26.Tsai KL, Yu X, Gopalan S, Chao TC, Zhang Y, Florens L, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544(7649):196–201. doi: 10.1038/nature21393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–5. doi: 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–95. doi: 10.1016/j.cell.2013.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta. 2013;1829(1):69–75. doi: 10.1016/j.bbagrm.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osman S, Mohammad E, Lidschreiber M, Stuetzer A, Bazso FL, Maier KC, et al. The Cdk8 kinase module regulates interaction of the mediator complex with RNA polymerase II. J Biol Chem. 2021;296:100734. doi: 10.1016/j.jbc.2021.100734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knuesel M, Meyer K, Bernecky C, Taatjes D. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–51. doi: 10.1101/gad.1767009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fant CB, Taatjes DJ. Regulatory functions of the Mediator kinases CDK8 and CDK19. Transcription. 2019;10(2):76–90. doi: 10.1080/21541264.2018.1556915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luyties O, Taatjes DJ. The Mediator kinase module: an interface between cell signaling and transcription. Trends Biochem Sci. 2022. doi: 10.1016/j.tibs.2022.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klatt F, Leitner A, Kim IV, Ho-Xuan H, Schneider EV, Langhammer F, et al. A precisely positioned MED12 activation helix stimulates CDK8 kinase activity. Proc Natl Acad Sci U S A. 2020;117(6):2894–905. doi: 10.1073/pnas.1917635117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol. 2013;20(5):611–9. doi: 10.1038/nsmb.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17(2):194–201. doi: 10.1038/nsmb.1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153(6):1327–39. doi: 10.1016/j.cell.2013.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526(7572):273–6. doi: 10.1038/nature14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging Views on the CTD Code. Genetics Research International. 2012;2012:347214. doi: 10.1155/2012/347214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–6. doi: 10.1038/35024111 [DOI] [PubMed] [Google Scholar]
- 41.Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139(4):757–69. doi: 10.1016/j.cell.2009.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, et al. CDK8 Kinase Phosphorylates Transcription Factor STAT1 to Selectively Regulate the Interferon Response. Immunity. 2013;38(2):250–62. doi: 10.1016/j.immuni.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29(3):650–61. doi: 10.1128/MCB.00993-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci U S A. 2010;107(25):11283–8. doi: 10.1073/pnas.0914215107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hengartner CJ, Myer VE, Liao S-M, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4 [DOI] [PubMed] [Google Scholar]
- 46.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95(5):717–28. doi: 10.1016/s0092-8674(00)81641-4 [DOI] [PubMed] [Google Scholar]
- 47.Feng D, Youn DY, Zhao X, Gao Y, Quinn WJ III, Xiaoli AM, et al. mTORC1 Down-Regulates Cyclin-Dependent Kinase 8 (CDK8) and Cyclin C (CycC). PLoS ONE. 2015;10(6):e0126240. doi: 10.1371/journal.pone.0126240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Feng D, Wang Q, Abdulla A, Xie XJ, Zhou J, et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J Clin Invest. 2012;122(7):2417–27. doi: 10.1172/JCI61462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poss ZC, Ebmeier CC, Odell AT, Tangpeerachaikul A, Lee T, Pelish HE, et al. Identification of Mediator Kinase Substrates in Human Cells using Cortistatin A and Quantitative Phosphoproteomics. Cell Rep. 2016;15(2):436–50. doi: 10.1016/j.celrep.2016.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galbraith MD, Andrysik Z, Pandey A, Hoh M, Bonner EA, Hill AA, et al. CDK8 Kinase Activity Promotes Glycolysis. Cell Rep. 2017;21(6):1495–506. doi: 10.1016/j.celrep.2017.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455(7212):552–6. doi: 10.1038/nature07310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zamudio AV, Dall’Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell. 2019;76(5):753–66 e6. doi: 10.1016/j.molcel.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen M, Liang J, Ji H, Yang Z, Altilia S, Hu B, et al. CDK8/19 Mediator kinases potentiate induction of transcription by NFkappaB. Proc Natl Acad Sci U S A. 2017;114(38):10208–13. doi: 10.1073/pnas.1710467114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birkenheuer CH, Brewster CD, Quackenbush SL, Rovnak J. Retroviral cyclin controls cyclin-dependent kinase 8-mediated transcription elongation and reinitiation. J Virol. 2015;89(10):5450–61. doi: 10.1128/JVI.00464-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brewster C, Birkenheuer C, Vogt M, Quackenbush S, Rovnak J. The retroviral cyclin of walleye dermal sarcoma virus binds cyclin-dependent kinases 3 and 8. Virology. 2011;409(2):299–307. doi: 10.1016/j.virol.2010.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rovnak J, Brewster CD, Quackenbush SL. Retroviral cyclin enhances cyclin-dependent kinase 8 activity. J Virol. 2012;86(10):5742–51. doi: 10.1128/JVI.07006-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, et al. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22(24):6494–504. doi: 10.1093/emboj/cdg619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vojnic E, Mourao A, Seizl M, Simon B, Wenzeck L, Lariviere L, et al. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat Struct Mol Biol. 2011;18(4):404–9. doi: 10.1038/nsmb.1997 [DOI] [PubMed] [Google Scholar]
- 59.Roupelieva M, Griffiths SJ, Kremmer E, Meisterernst M, Viejo-Borbolla A, Schulz T, et al. Kaposi’s sarcoma-associated herpesvirus Lana-1 is a major activator of the serum response element and mitogen-activated protein kinase pathways via interactions with the Mediator complex. J Gen Virol. 2010;91(Pt 5):1138–49. doi: 10.1099/vir.0.017715-0 [DOI] [PubMed] [Google Scholar]
- 60.Yang M, Hay J, Ruyechan WT. Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation. J Virol. 2008;82(24):12154–63. doi: 10.1128/JVI.01693-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyer TG, Martin MED, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–9. doi: 10.1038/20466 [DOI] [PubMed] [Google Scholar]
- 62.Songock WK, Kim SM, Bodily JM. The human papillomavirus E7 oncoprotein as a regulator of transcription. Virus Res. 2017;231:56–75. doi: 10.1016/j.virusres.2016.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rovnak J, Hronek BW, Ryan SO, Cai S, Quackenbush SL. An activation domain within the walleye dermal sarcoma virus retroviral cyclin protein is essential for inhibition of the viral promoter. Virology. 2005;342(2):240–51. doi: 10.1016/j.virol.2005.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniels CC, Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus Orf B functions through receptor for activated C kinase (RACK1) and protein kinase C. Virology. 2008;375:550–60. doi: 10.1016/j.virol.2008.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinparzer I, Sedlyarov V, Rubin JD, Eislmayr K, Galbraith MD, Levandowski CB, et al. Transcriptional Responses to IFN-gamma Require Mediator Kinase-Dependent Pause Release and Mechanistically Distinct CDK8 and CDK19 Functions. Mol Cell. 2019;76(3):485–99 e8. doi: 10.1016/j.molcel.2019.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leonard VH, Kohl A, Hart TJ, Elliott RM. Interaction of Bunyamwera Orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J Virol. 2006;80(19):9667–75. doi: 10.1128/JVI.00822-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas D, Blakqori G, Wagner V, Banholzer M, Kessler N, Elliott RM, et al. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J Biol Chem. 2004;279(30):31471–7. doi: 10.1074/jbc.M400938200 [DOI] [PubMed] [Google Scholar]
- 68.Pei J, Beri NR, Zou AJ, Hubel P, Dorando HK, Bergant V, et al. Nuclear-localized human respiratory syncytial virus NS1 protein modulates host gene transcription. Cell Rep. 2021;37(2):109803. doi: 10.1016/j.celrep.2021.109803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rice S, Kim SM, Rodriguez C, Songock W, Raikhy G, Lopez R, et al. Suppression of a Subset of Interferon-Induced Genes by Human Papillomavirus Type 16 E7 via a Cyclin Dependent Kinase 8-Dependent Mechanism. Viruses. 2020;12(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernandes-Siqueira LO, Zeidler JD, Sousa BG, Ferreira T, Da Poian AT. Anaplerotic Role of Glucose in the Oxidation of Endogenous Fatty Acids during Dengue Virus Infection. mSphere. 2018;3(1). doi: 10.1128/mSphere.00458-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fontaine KA, Sanchez EL, Camarda R, Lagunoff M. Dengue virus induces and requires glycolysis for optimal replication. J Virol. 2015;89(4):2358–66. doi: 10.1128/JVI.02309-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107(40):17345–50. doi: 10.1073/pnas.1010811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butler M, Chotiwan N, Brewster CD, DiLisio JE, Ackart DF, Podell B, et al. Cyclin-dependent kinases 8 and 19 regulate host cell metabolism during dengue virus serotype 2 infection. Viruses. 2020;12(6):654–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016;12(8):e1005841. doi: 10.1371/journal.ppat.1005841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petit MJ, Kenaston MW, Pham OH, Nagainis AA, Fishburn AT, Shah PS. Nuclear dengue virus NS5 antagonizes expression of PAF1-dependent immune response genes. PLoS Pathog. 2021;17(11):e1010100. doi: 10.1371/journal.ppat.1010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell. 2018;175(7):1931–45 e18. doi: 10.1016/j.cell.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]