Abstract
The IKK-NFκB complex is a key signaling node that facilitates activation of gene expression in response to extracellular signals. The kinase IKKβ and the transcription factor RELA have been targeted by covalent modifiers that bind to surface exposed cysteine residues. A common feature in well characterized covalent modifiers of RELA and IKKβ is the Michael acceptor containing α-methylene-γ-butyrolactone functionality. Through synthesis and evaluation of a focused set of α-methylene-γ-butyrolactone containing spirocyclic dimers (SpiDs) we identified SpiD3 as an anticancer agent with low nanomolar potency. Using cell-free and cell-based studies we show that SpiD3 is a covalent modifier that generates stable RELA containing high molecular weight complexes. SpiD3 inhibits TNFα-induced IκBα phosphorylation resulting in the blockade of RELA nuclear translocation. SpiD3 induces apoptosis, inhibits colony formation and migration of cancer cells. The NCI-60 cell line screen revealed that SpiD3 potently inhibits growth of leukemia cell lines, making it a suitable pretherapeutic lead for hematological malignancies.
Graphical Abstract
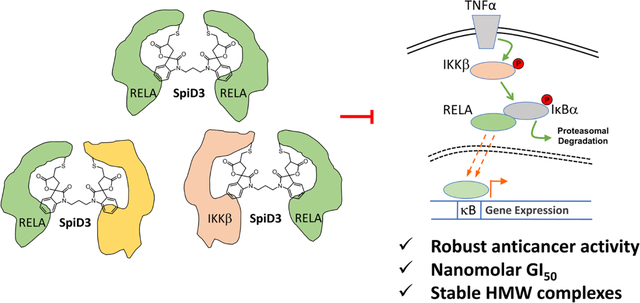
Nuclear factor kappa B (NFκB) is a transcription factor that plays a key role in immune response, host-immunity, inflammation, cell growth, and apoptosis.1 In resting cells, inhibitor of nuclear factor κB (IκBα) sequesters NFκB to the cytoplasm. The canonical NFκB pathway is activated by pro-inflammatory cytokines such as tumor necrosis factor alpha (TNFα). This leads to the phosphorylation of IκBα by the IκB kinase β (IKKβ), ubiquitination of p-IκBα by the E3-ligase (β-TrCP) and rapid degradation. The degradation of IκBα results in the nuclear translocation of NFκB and activation of gene expression.2–4 NFκB pathway is constitutively activated (ca) as evidenced by increased nuclear RELA levels in a variety of cancers including pancreatic, breast and ovarian cancers and has been shown to contribute to proliferation, tumor progression and chemoresistance.5–8 Immunohistochemistry (IHC) studies in surgically resected tumor samples, showed the presence of TNFα in the tumor microenvironment, which correlated with the presence of phosphorylated IKKβ in ~50% of tumor samples indicating the constitutive activation of the NFκB pathway.9 Consistently, expression of ca-IKKβ either in the absence of a tumor suppressor or the presence of oncogene accelerates tumor growth and dramatically reduced survival of mice.10, 11 This has led to a continued quest for small molecules and/or biologics that can block the sustained activation of the NF-κB pathway.
The IKK-NFκB complex is a ~900 KDa complex composed of several proteins that include kinases, transcription factors and scaffolding proteins.12, 13 Several small molecules have been reported to target different proteins in the IKK-NFκB complex. This includes, curcumin (an active ingredient of turmeric powder), NSAIDs such as aspirin, ibuprofen, sulindac, indomethacin and macrocyclic diaryl ether heptanoid natural products14 that are known to inhibit NFκB activation in cells. Bortezomib was approved by the FDA for multiple myeloma, a ubiquitin-proteasome inhibitor blocks the degradation of IκBα to prevent nuclear localization of RELA.15 Other strategies include antibodies against TNFα, inhibitors of IKKβ that block phosphorylation of IκBα and direct binders of RELA that block either the nuclear translocation of RELA or its ability to bind DNA.16
Sesquiterpene lactone (SL) natural products such as parthenolide (1), helenalin (2) and melampomagnolide B are also known modulators of NFκB signaling and have been studied extensively in many disease models (Figure 1).14, 17–22 For instance, numerous irreversible inhibitors including the SL natural products target surface exposed cysteine (SEC) residues in the NFκB pathway proteins. Well characterized SEC residues that are covalently modified are cys179 of IKKβ which results in the inhibition of IκBα phosphorylation and cys38 residue in RELA that inhibits NFκB mediated gene expression.17, 19, 23 A common feature among the natural products is the presence of α-methylene-γ-butyrolactone moiety which is critical for their activity against the NF-κB pathway proteins.19 Consequently, the α-methylene-γ-butyrolactone functionality was appended to several scaffolds and these hybrid molecules showed good anticancer activities.24–29 We reported the identification of a novel isatin derived spirocyclic core with the α-methylene-γ-butyrolactone (3, Figure 1) that covalently binds to IKKβ and RELA as a NFκB pathway inhibitor.27, 30 Compound 3 (analog 19) inhibited cell growth and induced apoptosis in a panel of cancer cell lines and with cisplatin synergistically inhibited tumor growth in an orthotopic ovarian cancer model without observable toxicity.27
Figure 1:
Previously reported anticancer sesquiterpene lactones and simplified helenalin derived NFκB Inhibitors.
The Crooks lab explored dimers of α-methylene-γ-butyrolactone containing natural products that could modify multiple SEC residues in glutathione-cysteine ligase enzyme complex as anticancer agents.31 Although the exact mechanism of action of these dimers was not characterized, they showed that the dimers of melampomagnolide B (4 & 5, Figure 1) exhibited potent antileukemic activity in primary AML cell lines. The Harki group reported simplified dimeric α-methylene-γ-butyrolactone containing Helenalin analogues that covalently modified SEC residues (cys38 and cys120) on RELA (6 & 7, Figure 1).23, 32 Recently, we showed that 3 binds to cys105 of RELA and cys46, 299, 412 and 524 of IKKβ. We also showed that 3, covalently modifies RELA and IKKβ in ovarian cancer cells. Since IKKβ and RELA are targeted by analog 3 and inspired by the above findings, we synthesized a spirocyclic dimer (SpiD7) of 3 that staples RELA to its binding partners resulting in the inhibition of TNFα induced nuclear translocation of RELA and NFκB mediated gene transcription.33 Here, we expand on the above findings by synthesis of a focused set of SpiD analogs and evaluated them for their anticancer activity. We report a 3-carbon linked dimer, SpiD3, as a low-nM anticancer agent that covalently modified RELA and IKKβ proteins to generate stable HMW homo- and hetero-complexes to block constitutive activation of the NFκB pathway.
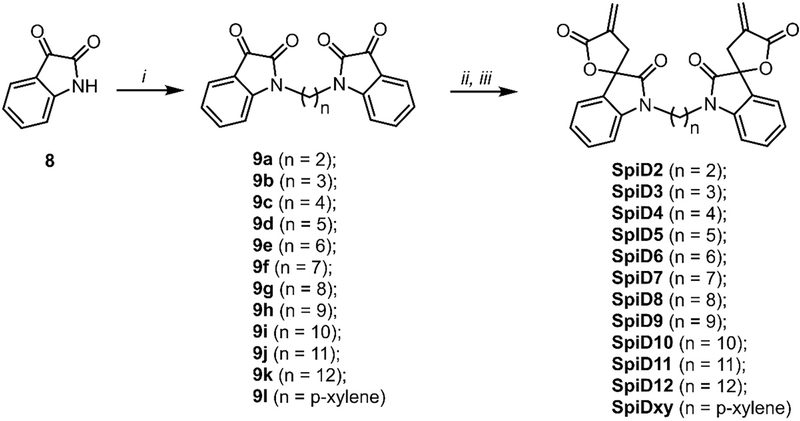
We previously showed that (a) analog 19 (3, Figure 1) covalently modified RELA and IKKβ (b) alkylation of the oxindole nitrogen atom in analog 19 was well tolerated, (c) the butyrolactone moiety in analog 19 was critical for its activity and (d) the reduction of exocyclic double bond in analog 19 resulted in complete loss of activity.27 Based on these preliminary observations, we synthesized dimers of analog 19 by alkylating the nitrogen atom of isatin (8) with a panel of dibromoalkanes in the presence of sodium hydride. These isatin dimers (9a - 9l) were subjected to an Indium metal catalyzed Barbier-type reaction to generate acyclic analogs.27, 30 An acid catalyzed ring closure of acyclic analogs yielded the desired spirocyclic dimers SpiD2 – SpiD12, & SpiDxy) with varying linkers (Scheme 1).
Scheme 1. Synthesis of dimeric spirocyclic based analogs.
(i). Dibromoalkane, NaH, DMF, RT, O/N (60–90%); (ii). Methyl 2-(bromomethyl)acrylate, In, THF: water, RT, 24h (40–70%); (iii). TsOH, DCM, RT, 24h (60–80%).
The focused set of SpiDs were screened for cancer cell growth inhibition (A2780, OVCAR5, and MiaPaCa2) in a three-day cell viability assay (Table 1). Among the panel of SpiDs, we found 3 carbon linked dimer (SpiD3) was the most potent inhibitor of cancer cell growth. SpiD3 was ~4–35- fold more potent (IC50) in inhibiting cancer cell growth when compared to the monomer analog 3. We also observed that linker length and composition affected growth inhibitory activities, with smaller linkers (2 – 7 carbon atoms) including the 6-atom constrained p-xylene linker showing better inhibition when compared to the longer linkers. Consistent with reported studies the reduced spirocyclic dimer (SpiD7-R) that lacks Michael acceptor did not inhibit growth (IC50 > 10 μM) of the cancer cell lines. This demonstrates that the cellular activity of the SpiDs like the monomer is probably due to the covalent modification of Cys residues.
Table 1.
Growth inhibition studies of α-methylene- γ-butyrolactone analogs in different cancer cell lines.
| Code | IC50 (μM) | |||
|---|---|---|---|---|
|
| ||||
| n | A2780 | OVCAR5 | MiaPaCa2 | |
|
| ||||
| Analog 19 | - | 0.99 ± 0.11 | 3.13 ± 0.55 | 2.05 ± 0.48 |
| SpiD2 | 2 | 0.39 ± 0.03 | 0.32 ± 0.02 | ND |
| SpiD3 | 3 | 0.23 ± 0.02 | 0.09 ± 0.01 | 0.13 ± 0.01 |
| SpiD4 | 4 | 0.36 ± 0.05 | 0.15 ± 0.03 | ND |
| SpiD5 | 5 | 0.27 ± 0.03 | 0.09 ± 0.01 | 0.20 ± 0.01 |
| SpiD6 | 6 | 0.41 ± 0.08 | 0.30 ± 0.02 | 0.23 ± 0.02 |
| SpiD7 | 7 | 0.41 ± 0.03 | 0.34 ± 0.06 | 0.26 ± 0.04 |
| SpiD8 | 8 | 0.60 ± 0.07 | 0.39 ± 0.05 | 0.74 ± 0.16 |
| SpiD9 | 9 | 1.08 ± 0.12 | 0.48 ± 0.06 | 1.22 ± 0.03 |
| SpiD10 | 10 | 0.49 ± 0.06 | 0.41 ± 0.06 | 0.56 ± 0.05 |
| SpiD11 | 11 | 0.77 ± 0.14 | 0.83 ± 0.12 | 2.03 ± 0.08 |
| SpiD12 | 12 | 0.50 ± 0.06 | 0.42 ± 0.55 | 1.5 ± 0.10 |
| SpiD-Xy | p-xylene | 0.31 ± 0.02 | 0.18 ± 0.01 | 0.27 ± 0.07 |
| SpiD7-R | 7 reduced | >10 | >10 | >10 |
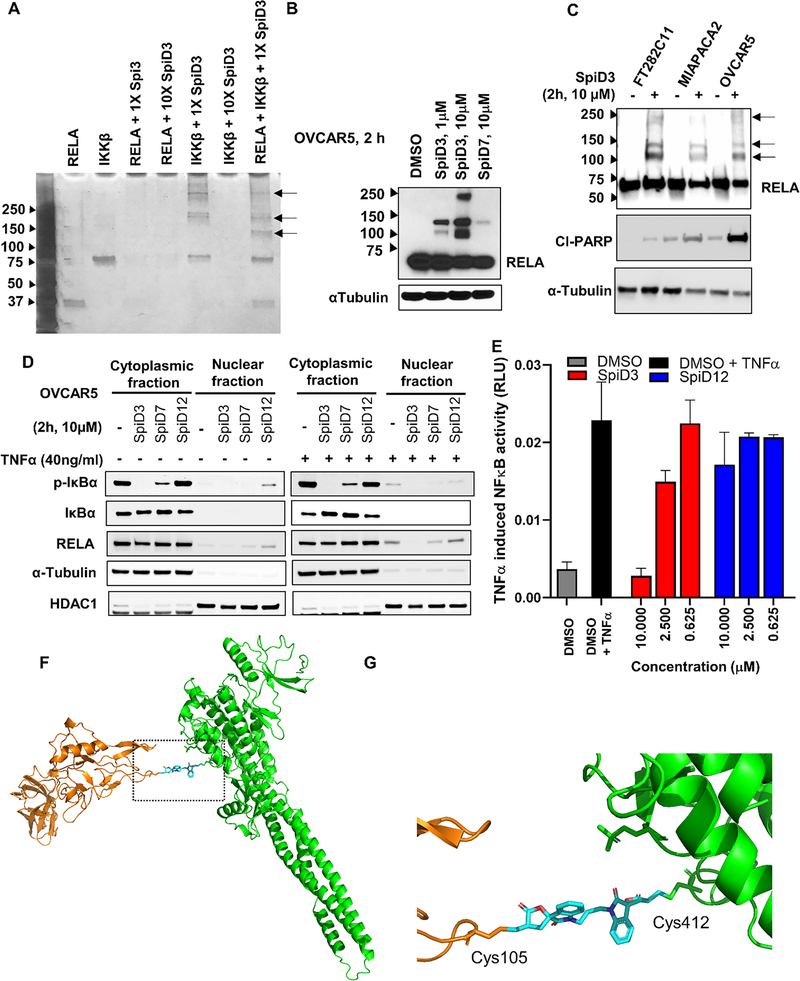
Since analog 19 covalently modifies RELA and IKKβ, we explored if the dimer SpiD3 crosslinks RELA and/or IKKβ proteins. To test this, we set up in vitro reactions with recombinant RELA and IKKβ wherein recombinant RELA and/or IKKβ proteins were incubated with equimolar or 10x concentrations of SpiD3. The resulting mixtures were separated by SDS-PAGE and subjected to silver staining to detect the recombinant proteins. We observed stable high molecular weight (HMW) bands in IKKβ:SpiD3 (1:1) and RELA:IKKβ:SpiD3 (1:1:1) lanes along with bands corresponding to unmodified IKKβ and RELA proteins (Figure 2A). We found no such HMW bands in the RELA:SpiD3 (1:1 or 1:10) lanes or in the IKKβ:SpiD3 (1:10) lane. Since we did not see any bands in the RELA:SpiD3 (1:1) lane but did see bands in the IKKβ:SpiD3 (1:1) lane (black arrows, Figure 2A), we conclude that the SEC residues in RELA are more reactive toward SpiD3 when compared to those in IKKβ. Analyses of the banding pattern in IKKβ: SpiD3 (1:1) and RELA:IKKβ:SpiD3 (1:1:1) lanes suggests formation of both homo- and hetero-dimers. Moreover, the presence of IKKβ disrupted SpiD3 mediated oligomerization of RELA protein by forming heterodimers of RELA-IKKβ complexes. This study shows that SpiD3 not only modifies both RELA and IKKβ, but also targets RELA with better efficiency, which is consistent with covalent docking energies of the monomer.33
Figure 2.
(A) Equimolar concentrations (7.72 μM) of the reactants were mixed and incubated for 24h at 4 °C. The mixture was subjected to SDS-PAGE and the gel subjected to silver staining silver staining of the in vitro RELA and IKKβ reaction with SpiD3. Black arrows indicate the HMW protein cross-link bands. (B) OVCAR5 cells were treated with and without SpiD3 and SpiD7 for 2 h. The lysates were subjected to WB analyses with antibodies against the indicated proteins. (C) WB analysis of panel of cell lines treated with and without 10 μM concentration of dimer SpiD3 for 2h. Black arrows indicate the HMW protein cross-link bands. (D) OVCAR5 cells were treated with 10μM dimers SpiD3, SpiD7 or SpiD12 for 2h and stimulated with and without 40ng/ml TNFα for 10 min post inhibitor treatment. The cytoplasmic and nuclear fractions were isolated, and lysates were subjected to western blotting. (E) Dose-response study with SpiD3 to assess inhibition of TNFα-induced RELA activation in a dual luciferase reporter assay (n=3, ±SD). (F) Proposed docking pose and (F) zoom in image showing crosslinking of SpiD3 to cys105 of RELA (pdb code 1IKN) and cys412 of IKKβ (pdb code 4KIK) respectively.
Next, to determine if linker length plays a role in the efficacy of the covalent modification resulting in the HMW complex formation, OVCAR5 cells were treated with SpiD3 or SpiD7 (7-carbon linker). The resulting lysates were subjected to western blot analyses and the membranes probed with RELA antibody. We found that SpiD3 treated cells showed higher levels of HMW bands as compared to SpiD7 (Figure 2B), indicating that the shorter linker more effectively modifies RELA. To assess if the covalent modification by SpiD3 translates to multiple cell lines, we treated an immortalized non-transformed fallopian tube epithelial cell (FT282C11), and two cancer cells (MiaPaCa2, and OVCAR5) with and without SpiD3. The lysates were subjected to western blot analyses and probed for RELA. Consistent with in vitro studies, we observed stable HMW bands in all 3 cell lines (Figure 2C) and observed modest reduction of basal RELA levels in the SpiD3 treated cancer cells (lane 4 & 6, Figure 2C) when compared to SpiD3 treated FT282C11 cells (lane 2, Figure 2C). We also observed robust PARP cleavage in SpiD3 treated cancer cells when compared to SpiD3 treated non-transformed immortalized cells. These results indicate that SpiD3 covalently modifies RELA in both normal and cancer cells but is likely inducing stronger degradation of RELA in cancer cells resulting in selective induction of cell death.
Next, we probed the functional consequence of SpiD3 mediated covalent modification of RELA on the canonical NFκB pathway. We monitored RELA localization in response to TNFα stimulation in cells treated with SpiD3, SpiD7 and SpiD12 (12-carbon linker). In its resting state, RELA is in complex with its endogenous inhibitor IκBα and is localized to the cytoplasm. Upon TNFα-stimulation, IκBα is phosphorylated, ubiquitinated and subsequently degraded. This enables RELA to translocate to the nucleus and activate gene expression.34, 35 We subjected two sets of OVCAR5 cells to DMSO, SpiD3 or SpiD7 or SpiD12. One set of cells were stimulated with TNFα while the other served as the control. Following a 10 min incubation, the lysates were fractionated and the cytoplasmic and nuclear fractions from the various treatments were subjected to western blot analyses (Figure 2D). Both in the TNFα stimulated and unstimulated samples, we observed that SpiD3 completely blocked phosphorylation of IκBα while 7-carbon linked SpiD7 partially inhibited IκBα phosphorylation and the 12-carbon linked SpiD12 did not inhibit IκBα phosphorylation. Consistently the nuclear RELA levels mirrored the effects observed with phosphorylated IκBα wherein, SpiD3 treatment potently inhibited nuclear localization of RELA while SpiD7 partially inhibited nuclear localization of RELA and SpiD12 had no effect on the nuclear localization of RELA. This demonstrates that the linker length plays a key role in the ability of SpiDs to inhibit TNFα induced IκBα phosphorylation and the levels of nuclear RELA correlates with the growth inhibitory effects of the SpiDs tested.
To assess the effects of TNFα-induced nuclear translocation of RELA on NFκB mediated gene expression, we conducted a dose-response study using a TNFα-induced NFκB reporter assay with SpiD3 and SpiD12 (Figure 2E). The results show that SpiD3 inhibited TNFα-induced NFκB activity in a dose-dependent manner while SpiD12 did not. Together, our studies demonstrate that covalent modification of RELA and IKKβ by SpiD3 blocks IκBα phosphorylation resulting in the inhibition of RELA nuclear localization and NFκB mediated gene expression. Covalent docking using Schrodinger GLIDE identified pockets adjacent to Cys105 on RELA and Cys412 on IKKβ as the most accessible for analog 19 binding.33 Figure 2F summarizes our model for SpiD3 mediated covalent modification of RELA and IKKβ to generate the observed stable HMW hetero complex.
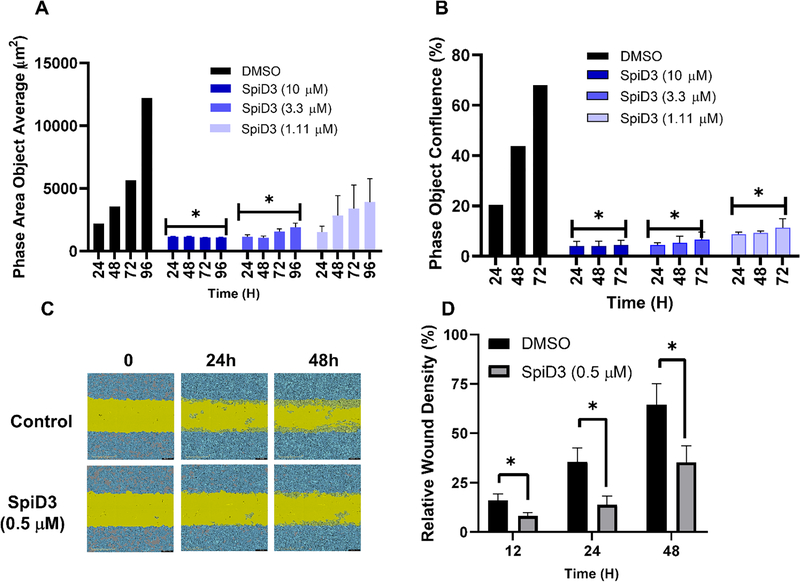
The ability of a cell to undergo unlimited cell division is routinely assessed in vitro using clonogenic assay.36 Here we used an Incucyte® live cell imaging study to assess the ability of SpiD3 to inhibit colony formation. Ovarian cancer cells (A2780) were sparsely plated and allowed to grow in the presence or absence of SpiD3 for 4 days under continues monitoring. The colonies were quantified using Incucyte® S3 live-cell imaging and tracking software (Figure 3A). When compared to DMSO treated wells, SpiD3 treatment showed a dose-dependent decrease in the number of colonies across different time points (Figure 3A). Since SpiD3 is a covalent modifier that forms irreversible C-S bonds through the α-methylene-γ-butyrolactone moiety, we also evaluated the ability of SpiD3 to inhibit cell proliferation in a “no inhibitor, washout” assay. Briefly, A2780 cells were treated with increasing concentrations of SpiD3, following a 6h incubation, media containing SpiD3 was removed, and replaced with fresh media. Cells were then allowed to proliferate without inhibitor for 72h (Figure 3B). SpiD3 treated wells showed robust inhibition of cell growth across all treated wells suggesting that the effects induced by SpiD3 occur within the first 6h of treatment. Since TNFα contributes to migration, invasion, and angiogenesis,37 we performed a wound-healing scratch assay to assess the effect of SpiD3 on the ability of cells to migrate.38 A2780 cells were treated with DMSO or 500nM of SpiD3 and imaged at 0, 12, 24, and 48h (Figure 3C & D). Quantification of wound healing showed that SpiD3 inhibited migration of A2780 cells by ~50% when compared to cells in DMSO treated wells.
Figure 3:
(A) SpiD3 inhibited the colony formation in A2780 cells. (B) A2780 cells were treated with SpiD3 for 6h at difference concentrations. Cell growth medium containing SpiD3 was then removed (washout) and cells were allowed to grow over three days. (C) SpiD3 disrupts migration of A2780 cells. Wound gap images taken during the 24h and 48h incubation of HCT116 cells with DMSO or 500 nM SpiD3. (D) Quantification of % wound closure after treatment of HCT116 cells with DMSO of 500 nM SpiD3.
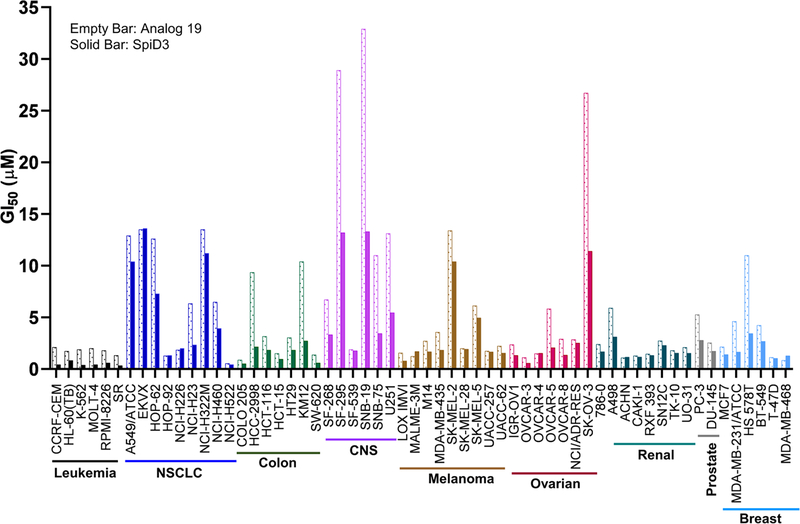
Next, to assess the effects of SpiD3 across a panel of cell lines, we submitted analog 19 and SpiD3 to NCI for evaluation in the NCI-60 human cancer cell line screening. SpiD3 was screened at five concentrations (10-fold dilutions) and its ability to inhibit cancer cell growth (GI50) is summarized in Figure 4. SpiD3 inhibited cancer cell line growth with sub- to low-μM potencies with the Leukemia (GI50 ~ 0.38 – 0.83 μM) cell lines being the most sensitive and CNS (GI50 ~ 1.76 – 13.3 μM) cell lines were the least sensitive. When compared to the monomer analog 19, the dimer SpiD3 showed improved growth inhibitory effects in >90% cell lines (Figure 4 and Supplementary Figure S1 – S6). A similar screen was also performed by Janganati et al with the dimers of Melampomagnolide B.31 Although they didn’t compare GI50 values of monomer vs. dimer, they showed that dimer 7f exhibited promising activity against all cancer cell lines. Among the leukemia lines SpiD3 (AveGI50 = 0.49 ± 0.19 μM) and 7f (AveGI50 = 0.29 ± 0.04 μM) have comparable activities.
Figure 4:
Growth Inhibition (GI50) for SpiD3 and monomeric analog 19 against a panel of 60 Human Cancer Cell Lines. GI50 is the drug concentration resulting in a 50% reduction in the net protein increase (as measured by SRB staining) in control cells during the drug incubation.
The goal of the NCI 60-cell line panel screen was to discover cell-type specific agents with clinical activity for the corresponding solid tumors. Analyses of the data showed that the correlation between histology and clinical activity was low but the drug sensitivity correlated with the molecular target expression.39 We extracted the protein levels of RELA, IKKβ and IκBα, which are the three proteins that we showed were perturbed by SpiD3 from cell lines that were common between the NCI-60-cell line panel and the reported cancer cell line proteomics encyclopedia.40 Plots between the growth inhibitory effects of SpiD3 with the above protein levels showed that IκBα levels exhibited the highest correlation (R = 0.28) with the GI50 values (Supplementary Table S1). This suggests that inhibition of IκBα phosphorylation correlates with the growth inhibitory effects of SpiD3.
In conclusion, we report the synthesis of isatin derived spirocyclic α-methylene-γ-butyrolactone dimers and evaluated them for their anticancer activities. The screen identified a 3-carbon linked dimer, SpiD3, as an anticancer agent with low nanomolar potency. SpiD3 covalently modified RELA and IKKβ proteins to generate stable HMW homo- and hetero-complexes. When compared to non-transformed immortalized cells, SpiD3 induced formation of RELA containing HMW complexes and robust PARP cleavage in cancer cells. We also showed that linker length plays a key role in the ability of SpiDs to inhibit TNFα-induced nuclear localization of RELA which correlates with the growth inhibitory effects. SpiD3 inhibited growth, colony formation, and migration of cancer cells. Profiling SpiD3 in the NCI-60 cell line panel showed that leukemia cell lines were most sensitive. In summary, our data shows that SpiD3 exhibits anticancer effects by covalent modification of proteins in the IKK-NFκB complex.
Supplementary Material
Acknowledgements:
This work was supported in part by NIH grants (CA197999, CA251151, CA260749 and CA036727), NRI, LB506 and FPBCC pilot grant. S. Ko. was supported by UNMC fellowship. Research resources at UNMC are supported in part by the Nebraska Research Initiative. Biochemistry research at SDSU is supported in part by the California Metabolic Research Foundation.
Footnotes
Supplementary data
Supplementary data associated with this article can be found in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Oeckinghaus A; Ghosh S The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 2009, 1, a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F; Xia Y; Parker AS; Verma IM IKK biology. Immunol Rev 2012, 246, 239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiDonato JA; Hayakawa M; Rothwarf DM; Zandi E; Karin M A cytokine-responsive IkB kinase that activates the transcription factor NF-kB. Nature 1997, 388, 549–554. [DOI] [PubMed] [Google Scholar]
- 4.DiDonato J; Mercurio F; ROSETTE C; WU-LI J; SUYANG H; GHOSH S; KARIN M Mapping of the Inducible IkB Phosphorylation Sites That Signal Its Ubiquitination and Degradation. Mol Cell Biol 1996, 16, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabhu L; Mundade R; Korc M; Loehrer PJ; Lu T Critical role of NF-kappaB in pancreatic cancer. Oncotarget 2014, 5, 10969–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sau A; Lau R; Cabrita MA; Nolan E; Crooks PA; Visvader JE; Pratt MA Persistent Activation of NF-kappaB in BRCA1-Deficient Mammary Progenitors Drives Aberrant Proliferation and Accumulation of DNA Damage. Cell Stem Cell 2016, 19, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvero AB Recent insights into the role of NF-κB in ovarian carcinogenesis. genome medicine 2010, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Y; Shen S; Verma IM NF-kappaB, an active player in human cancers. Cancer Immunol Res 2014, 2, 823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee DF; Kuo HP; Chen CT; Hsu JM; Chou CK; Wei Y; Sun HL; Li LY; Ping B; Huang WC; He X; Hung JY; Lai CC; Ding Q; Su JL; Yang JY; Sahin AA; Hortobagyi GN; Tsai FJ; Tsai CH; Hung MC IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 2007, 130, 440–55. [DOI] [PubMed] [Google Scholar]
- 10.Shaked H; Hofseth LJ; Chumanevich A; Chumanevich AA; Wang J; Wang Y; Taniguchi K; Guma M; Shenouda S; Clevers H; Harris CC; Karin M Chronic epithelial NF-kappaB activation accelerates APC loss and intestinal tumor initiation through iNOS upregulation. Proc Natl Acad Sci U S A 2012, 109, 14007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naramura M; Natarajan A Mouse Pancreatic Tumor Model Independent of Tumor Suppressor Gene Inactivation. Pancreas 2018, 47, e27–e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiDonato JA; Hayakawa M; Rothwarf DM; Zandi E; Karin M A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature 1997, 388, 548–54. [DOI] [PubMed] [Google Scholar]
- 13.Mercurio F; Zhu H; Murray BW; Shevchenko A; Bennett BL; Li J; Young DB; Barbosa M; Mann M; Manning A; Rao A IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 1997, 278, 860–6. [DOI] [PubMed] [Google Scholar]
- 14.Bryant VC; Kishore Kumar GD; Nyong AM; Natarajan A Synthesis and evaluation of macrocyclic diarylether heptanoid natural products and their analogs. Bioorg Med Chem Lett 2012, 22, 245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane RC; Bross PF; Farrell AT; Pazdur R Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist 2003, 8, 508–13. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore TD; Herscovitch M Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene 2006, 25, 6887–99. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto G; Namekawa J; Muta M; Nakamura T; Bando H; Tohyama K; Toi M; Umezawa K Targeting of nuclear factor kappaB Pathways by dehydroxymethylepoxyquinomicin, a novel inhibitor of breast carcinomas: antitumor and antiangiogenic potential in vivo. Clin Cancer Res 2005, 11, 1287–93. [PubMed] [Google Scholar]
- 18.Govindachari TRJ, S. B; Kamat VN Structure of parthenolide. Tetrahedron 1965, 21, 1509–1519. [Google Scholar]
- 19.Kwok BH; Koh B; Ndubuisi MI; Elofsson M; Crews CM The anti-inflammatory natural product parthenolide from the medicinal herb Feverfew directly binds to and inhibits IkappaB kinase. Chem Biol 2001, 8, 759–66. [DOI] [PubMed] [Google Scholar]
- 20.Liang MC; Bardhan S; Pace EA; Rosman D; Beutler JA; Porco JA Jr.; Gilmore TD Inhibition of transcription factor NF-kappaB signaling proteins IKKbeta and p65 through specific cysteine residues by epoxyquinone A monomer: correlation with its anti-cancer cell growth activity. Biochem Pharmacol 2006, 71, 634–45. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M; Horie R; Takeiri M; Kozawa I; Umezawa K Inactivation of NFkappaB components by covalent binding of (−)-dehydroxymethylepoxyquinomicin to specific cysteine residues. J Med Chem 2008, 51, 5780–8. [DOI] [PubMed] [Google Scholar]
- 22.Kitson RR; Millemaggi A; Taylor RJ The renaissance of alpha-methylenegamma-butyrolactones: new synthetic approaches. Angew Chem Int Ed Engl 2009, 48, 9426–51. [DOI] [PubMed] [Google Scholar]
- 23.Widen JC; Kempema AM; Villalta PW; Harki DA Targeting NF-kappaB p65 with a Helenalin Inspired Bis-electrophile. ACS Chem Biol 2017, 12, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraldi PG; Del Carmen Nunez M; Tabrizi MA; De Clercq E; Balzarini J; Bermejo J; Estevez F; Romagnoli R Design, synthesis, and biological evaluation of hybrid molecules containing alpha-methylene-gamma-butyrolactones and polypyrrole minor groove binders. J Med Chem 2004, 47, 2877–86. [DOI] [PubMed] [Google Scholar]
- 25.Ropp S; Guy J; Berl V; Bischoff P; Lepoittevin JP Synthesis and photocytotoxic activity of new alpha-methylene-gamma-butyrolactone-psoralen heterodimers. Bioorg Med Chem 2004, 12, 3619–25. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran PV; Pratihar D; Nair HN; Walters M; Smith S; Yip-Schneider MT; Wu H; Schmidt CM Tailored alpha-methylene-gamma-butyrolactones and their effects on growth suppression in pancreatic carcinoma cells. Bioorg Med Chem Lett 2010, 20, 6620–3. [DOI] [PubMed] [Google Scholar]
- 27.Rana S; Blowers EC; Tebbe C; Contreras JI; Radhakrishnan P; Kizhake S; Zhou T; Rajule RN; Arnst JL; Munkarah AR; Rattan R; Natarajan A Isatin Derived Spirocyclic Analogues with alpha-Methylene-gamma-butyrolactone as Anticancer Agents: A Structure-Activity Relationship Study. J Med Chem 2016, 59, 5121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran PV; Nicponski DR; Nair HN; Helppi MA; Gagare PD; Schmidt CM; Yip-Schneider MT Synthetic alpha-(aminomethyl)-gamma-butyrolactones and their anti-pancreatic cancer activities. Bioorg Med Chem Lett 2013, 23, 6911–4. [DOI] [PubMed] [Google Scholar]
- 29.Rana S; Kour S; Sonawane YA; Robb CM; Contreras JI; Kizhake S; Zahid M; Karpf AR; Natarajan A Symbiotic prodrugs (SymProDs) dual targeting of NFkappaB and CDK. Chem Biol Drug Des 2020, 96, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rana S; Natarajan A Face selective reduction of the exocyclic double bond in isatin derived spirocyclic lactones. Org Biomol Chem 2013, 11, 244–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janganati V; Ponder J; Jordan CT; Borrelli MJ; Penthala NR; Crooks PA Dimers of Melampomagnolide B Exhibit Potent Anticancer Activity against Hematological and Solid Tumor Cells. J Med Chem 2015, 58, 8896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Widen JC; Kempema AM; Baur JW; Skopec HM; Edwards JT; Brown TJ; Brown DA; Meece FA; Harki DA Helenalin Analogues Targeting NF-kappaB p65: Thiol Reactivity and Cellular Potency Studies of Varied Electrophiles. ChemMedChem 2018, 13, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit Kour SR, Smitha Kizhake, Dragana Lagundžin, David Klinkebiel, Jayapal Reddy Mallareddy, Tom Huxford, Nicholas T. Woods, and Amarnath Natarajan. Stapling proteins in the RELA complex inhibits TNFa-induced nuclear translocation of RELA. RSC Chemical Biology 2022, 3, 32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldwin AS Jr. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 1996, 14, 649–83. [DOI] [PubMed] [Google Scholar]
- 35.Baeuerle PA; Baltimore D NF-kappa B: ten years after. Cell 1996, 87, 13–20. [DOI] [PubMed] [Google Scholar]
- 36.Franken NA; Rodermond HM; Stap J; Haveman J; van Bree C Clonogenic assay of cells in vitro. Nat Protoc 2006, 1, 2315–9. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y; Zhou BP TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer 2010, 102, 639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robb CM; Kour S; Contreras JI; Agarwal E; Barger CJ; Rana S; Sonawane Y; Neilsen BK; Taylor M; Kizhake S; Thakare RN; Chowdhury S; Wang J; Black JD; Hollingsworth MA; Brattain MG; Natarajan A Characterization of CDK(5) inhibitor, 20–223 (aka CP668863) for colorectal cancer therapy. Oncotarget 2018, 9, 5216–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monks A; Scudiero DA; Johnson GS; Paull KD; Sausville EA The NCI anti-cancer drug screen: a smart screen to identify effectors of novel targets. Anticancer Drug Des 1997, 12, 533–41. [PubMed] [Google Scholar]
- 40.Nusinow DP; Szpyt J; Ghandi M; Rose CM; McDonald ER 3rd; Kalocsay M; Jane-Valbuena J; Gelfand E; Schweppe DK; Jedrychowski M; Golji J; Porter DA; Rejtar T; Wang YK; Kryukov GV; Stegmeier F; Erickson BK; Garraway LA; Sellers WR; Gygi SP Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.