Abstract
Abstract
Dilated cardiomyopathy (DCM) is the third most common cause of heart failure. The multidisciplinary nature of testing — involving genetics, imaging, or cardiovascular techniques — makes its diagnosis challenging. Novel and reliable biomarkers are needed for early identification and tailored personalized management. Peripheral circular RNAs (circRNAs), a leading research topic, remain mostly unexplored in DCM. We aimed to assess whether peripheral circRNAs are expressed differentially among etiology-based DCM. The study was based on a case–control multicentric study. We enrolled 130 subjects: healthy controls (n = 20), idiopathic DCM (n = 30), ischemic DCM (n = 20), and familial DCM patients which included pathogen variants of (i) LMNA gene (n = 30) and (ii) BCL2-associated athanogene 3 (BAG3) gene (n = 30). Differentially expressed circRNAs were analyzed in plasma samples by quantitative RT-PCR and correlated to relevant systolic and diastolic parameters. The pathophysiological implications were explored through bioinformatics tools. Four circRNAs were overexpressed compared to controls: hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239 in LMNA-related DCM and hsa_circ_0089762 in the ischemic DCM cohort. The obtained areas under the curve confirm the discriminative capacity of circRNAs. The circRNAs correlated with some diastolic and systolic echocardiographic parameters with notable diagnostic potential in DCM. Circulating circRNAs may be helpful for the etiology-based diagnosis of DCM as a non-invasive biomarker.
Key messages
The limitations of cardiac diagnostic imaging and the absence of a robust biomarker reveal the need for a diagnostic tool for dilated cardiomyopathy (DCM).
The circular RNA (circRNA) expression pattern is paramount for categorizing the DCM etiologies.
Our peripheral circRNAs fingerprint discriminates between various among etiology-based DCM and correlates with some echocardiographic parameters.
We provide a potential non-invasive biomarker for the etiology-based diagnosis of LMNA-related DCM and ischemic DCM.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00109-021-02119-6.
Keywords: Circulating circular RNA, Ischemic-dilated cardiomyopathy, Lamin A/C-dilated cardiomyopathy
Introduction
Heart failure is a global pandemic affecting more than 25 million people worldwide, with a continuously increasing prevalence [1]. One of the major causes of heart failure is dilated cardiomyopathy (DCM), characterized by chamber enlargement and contractile dysfunction of the left ventricle (LV) [2]. Several etiologies are included in the DCM common pathway. Ischemic cardiomyopathy is more common than non-ischemic (59% compared with 41%) [2]. Among non-ischemic cardiomyopathy, up to 35% of idiopathic DCM may have a family history [2, 3]. Pathogenic alterations in the gene encoding nuclear lamin A and C proteins-lamin A/C (LMNA) explain 5–10% of familial DCM cases.
DCM is a heterogeneous entity that has different outcomes and may require diverse therapies [4]. Notably, ischemic and familial DCM are major groups with life-threatening arrhythmias [3]. LMNA-related DCM presents highly aggressive outcomes and lethal ventricular arrhythmias [5]. Male sex, LV ejection fraction (LVEF) lower than 50%, and non-missense mutations are independent predictors of adverse outcome [6]. Thus, the identification of DCM etiology may help clinicians to stratify patients at risk of fatal events. However, the diagnosis process to reach DCM etiology involves several clinical steps. Multidisciplinary teams, imaging tests, the high cost of genetic testing, and its low efficiency make DCM etiologic diagnosis challenging. A precise, accessible biomarker that supports this process is required to improve diagnosis and early identification of asymptomatic cases. This would facilitate the adoption of tailored management.
Non-coding RNAs have pivotal roles in regulating the network that governs the physiology and pathology of cardiovascular diseases [7]. To date, microRNAs (miRNAs) have been considered a more relevant biomarker candidate due to the complexity of circular RNA (circRNA) assessment in human screening [8]. However, circRNAs also have thought-provoking features. The advantages of circRNAs are their cell type, tissue, and developmental stage specificity. Furthermore, they are independently regulated and more stable than lineal RNA, and they are gathered in cells and human body fluids [8]. CircRNAs modulate gene expression by sponging miRNAs, interacting with RNA-binding proteins (RBPs), and competing with canonical splicing of their pre-mRNA precursor [9]. Research reporting circRNAs as an effective diagnostic and therapeutic biomarker in many diseases has grown exponentially in the last decade. Nevertheless, the potential of using this easy-to-monitor and highly stable marker for stratifying DCM etiologies remains unexplored. Additionally, the experimental and computational analyses of these molecular cross-regulations will propel new insights on DCM [8].
The present study aimed to identify differentially expressed circRNAs in the plasma of patients with DCM of various etiologies such as familial, idiopathic, or ischemic.
Material and methods
Study design
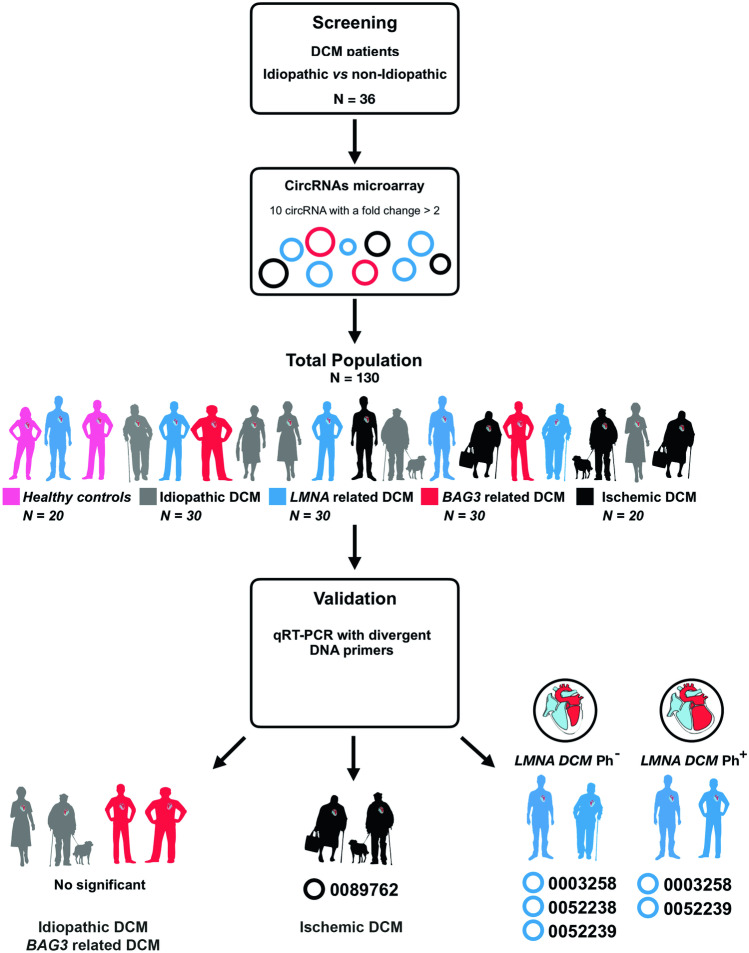
The study was based on a case–control multicentric study. Patient samples and the dataset were collected from several centers (Puerta del Mar University Hospital, Cádiz; Cruz Roja Hospital, Madrid; and Virgen de la Victoria University Hospital, Málaga, Spain). We enrolled 130 subjects distributed in five study groups: healthy controls (n = 20), idiopathic DCM (n = 30), ischemic DCM (n = 20), and familial DCM patients. The carriers of rare pathogenic variants included were (i) LMNA gene (n = 30) and (ii) BCL2-associated athanogene 3 (BAG3) gene (n = 30) (Fig. 1).
Fig. 1.
Flowchart of the study design strategy. This figure illustrates the experimental workflow of the study including screening, validation, and peripheral circRNAs overexpressed for the LMNAPh−, LMNAPh+, and ischemic DCM cohort. Abbreviations: BAG3, BCL2-associated athanogene 3; DCM, dilated cardiomyopathy; lamin A/C; LMNAPh−, LMNA carrier of the pathogenic variant; LMNAPh+, LMNA carrier phenotype positive; LVEF, left ventricle ejection fraction
DCM etiology was determined by three independent clinical cardiologists, who are experts in cardiomyopathies. DCM was defined as either LVEF levels below 50% and/or LV end-diastolic diameter larger than 56 mm [10]. BAG3 and LMNA participants were confirmed genetically and fulfilled the diagnostic clinical criteria for familial DCM [11]. The LMNA cohort was subclassified as a carrier of the pathogenic variant, phenotypically negative (LMNAPh−) and genetically and phenotypically positive (LMNAPh+) as previously described [11]. Genetic etiology was ruled out in all idiopathic DCM patients. Ischemic DCM was diagnosed if a precedent of acute myocardial infarction or coronary artery disease was shown, which developed LV remodeling and dysfunction [10]. A transthoracic echocardiography protocol was performed as described previously [11, 12]. The information included anthropometric, clinical, therapeutic, electrocardiographic, and echocardiographic data from electronic medical records (Table 1).
Table 1.
Study population: anthropometric, clinical, and echocardiographic variables
| Variable | Healthy control (N = 20) | Idiopathic (N = 30) | LMNAPh− (N = 12) | LMNAPh+ (N = 18) | BAG3 (N = 30) | Ischemic (N = 20) |
|---|---|---|---|---|---|---|
| Age (years) | 42.0 ± 11.0 | 63.7 ± 8.2 | 40.6 ± 6.9 | 38.7 ± 15.0 | 42.2 ± 14.8 | 71.1 ± 8.5 |
| Sex (male) | 55% | 70% | 23.1% | 42.9% | 68.4% | 72.2% |
| BMI (kg/m2) | 25.1 ± 3.3 | 26.7 ± 2.6 | 25.4 ± 2.1 | 23.6 ± 3.9 | 27.9 ± 4.9 | 28.8 ± 4.9 |
| Heart rate (bpm) | 65.7 ± 11.9 | 71 ± 13.9 | 65.7 ± 5.9 | 64.3 ± 9.9 | 73 ± 10 | 64.6 ± 16.8 |
| Smoker | 0% | 60% | 57.1% | 30.8% | 26.3% | 22.2% |
| SBP (mm Hg) | 114.5 ± 8.7 | 113.1 ± 11.9 | 128.4 ± 15.9 | 123.2 ± 20.9 | 128.1 ± 13.3 | 124.3 ± 12.7 |
| DBP (mm Hg) | 73.5 ± 8.5 | 73.1 ± 7.1 | 81.8 ± 6.1 | 76.7 ± 17.9 | 81.1 ± 7.8 | 72.2 ± 8.6 |
| LVEF (%) | 68.8 ± 6.0 | 30.5 ± 10.2 | 44.5 ± 5.0 | 61.0 ± 5.9 | 49.5 ± 11.9 | 34.7 ± 7.5 |
| LVEDD (mm) | 47.7 ± 4.8 | 63.0 ± 3.8 | 58.0 ± 3.4 | 49.2 ± 12.6 | 55.6 ± 7.5 | 58.6 ± 4.8 |
| LVESD (mm) | 30.0 ± 6.9 | 48.1 ± 16.8 | 43.8 ± 3.1 | 30.7 ± 6.8 | 40.4 ± 9.3 | 44.1 ± 13.2 |
| LA volume (mL/m2) | 17.4 ± 4.3 | 71.1 ± 25.0 | 49.3 ± 12.4 | 41.0 ± 15.5 | 68.2 ± 25.8 | 62.1 ± 19.6 |
| LAD (mm) | 35.1 ± 5.4 | 45.2 ± 9.1 | 40.8 ± 4.3 | 33.8 ± 6.6 | 37.6 ± 6.5 | 40.8 ± 6.1 |
| RV (mm) | 28.6 ± 3.5 | 39.7 ± 6.5 | 31.7 ± 1.9 | 28.8 ± 5.2 | 32.1 ± 7.6 | 31.4 ± 6.9 |
| TAPSE | 22.2 ± 2.7 | 18.2 ± 6.4 | 21.6 ± 3.6 | 21.3 ± 3.5 | 21.1 ± 5.4 | 18.8 ± 3.9 |
| MAPSE | 18.1 ± 1.6 | 9.6 ± 2.7 | 12.1 ± 3.1 | 16.0 ± 2.6 | 12.3 ± 3.2 | 10.6 ± 2.1 |
| E (cm/s) | 0.7 ± 0.2 | 0.7 ± 0.2 | 0.8 ± 0.1 | 0.8 ± 0.2 | 0.8 ± 0.3 | 0.8 ± 0.2 |
| A (cm/s) | 0.6 ± 0.1 | 0.8 ± 0.3 | 0.7 ± 0.3 | 0.5 ± 0.2 | 0.6 ± 0.2 | 0.8 ± 0.3 |
| S’sTDI (cm/s) | 0.08 ± 0.01 | 0.06 ± 0.06 | 0.06 ± 0.01 | 0.08 ± 0.02 | 0.08 ± 0.01 | 0.05 ± 0.01 |
| E’s TDI (cm/s) | 0.09 ± 0.03 | 0.05 ± 0.05 | 0.07 ± 0.02 | 0.10 ± 0.04 | 0.09 ± 0.04 | 0.05 ± 0.01 |
| A’s TDI (cm/s) | 0.10 ± 0.03 | 0.06 ± 0.02 | 0.11 ± 0.02 | 0.09 ± 0.04 | 0.11 ± 0.03 | 0.07 ± 0.03 |
| E/E’ ratio | 7.7 ± 2.1 | 16.3 ± 8.5 | 10.2 ± 3.0 | 7.9 ± 2.2 | 6.3 ± 1.5 | 15.0 ± 6.4 |
| NYHA functional class (II-III) | 0% | 10% | 14.3% | 15.4% | 10.5% | 11.1% |
All values are expressed as mean ± SEM
A atrial systolic transmitral flow wave, A’s TDI atrial septal mitral annular velocity, BAG3 BCL2-associated athanogene 3, BMI body mass index, DBP diastolic blood pressure, DCM dilated cardiomyopathy, E early diastolic transmitral flow wave, E′ early diastolic mitral annular velocity, LA left atrial, LAD left atrial dimension, LMNA lamin A/C, LMNAPh− LMNA carrier of the pathogenic variant, LMNAPh+ LMNA carrier phenotypically positive, LVEDD left ventricular end-diastolic dimension, LVEF left ventricle ejection fraction, LVESD left ventricle end-systolic dimension, MAPSE mitral annular plane systolic excursion, NYHA New York Heart Association classification, RV right ventricle, S’ positive systolic wave, SBP systolic blood pressure, TAPSE tricuspid annular plane systolic excursion, TDI tissue Doppler imaging
Ethics
The study protocol was approved by the Andalusian Biomedical Research Ethics committee. The study was performed in full compliance with the Declaration of Helsinki. All participants provided written informed consent.
Genetic analysis
Genetic analysis was performed as previously described [11]. DNA isolation was undertaken using Chemagic MSM I from whole blood (Chemagic Human Blood). DNA integrity was assessed on 0.8% agarose gel and the quality ratios of absorbance were accomplished using spectrophotometric measurements. dsDNA concentration was determined using fluorometry integrity (Qubit, Life Technologies) and corroborated on 0.8% agarose gel.
Blood collection
Ten milliliters of peripheral blood was collected in K2-ethylenediaminetetraacetic acid tubes (BD) after 10 h overnight fasting. None of the patients was under heparin therapy. The blood was processed within 4 h after isolation, centrifuged (1500 g, 15 min, 4 °C), and the plasma layer was aliquoted and stored at − 80ºC until further analysis.
Microarray analysis
A screening study was carried out using the Arraystar Human Circular RNA Microarray V2.0 (Arraystar, Inc.). This platform analyzed 36 samples of idiopathic and non-idiopathic DCM subjects. Total RNAs from each sample were obtained using the Arraystar’s standard protocols (Arraystar, Inc.). The enriched circRNAs were amplified and transcribed into fluorescent cDNA using a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labelled cDNAs were hybridized onto the Arraystar Human circRNA Array V2.0 (Arraystar, Inc.). Once the slides had been washed, they were scanned by the Agilent Scanner G2505C.
RNA isolation and quantitative reverse transcriptase-polymerase chain reaction
Total RNA was isolated from 200 µL of plasma using a miRNeasy Serum/Plasma Kit (Qiagen). RNA was eluted with 20 µL of RNase-free H2O and stored at − 80 °C. For the circRNA quantification, circulating RNA preparations were reverse transcribed with a first-strand cDNA synthesis kit (Nzytech, Portugal) using a random primer approach and following the manufacturer’s instructions. Previous to reverse transcription, samples were spiked with MS2 RNA (Sigma-Aldrich, Germany), which was used as an internal normalizer. Quantification of selected circRNAs was performed by qRT-PCR using divergent DNA primers designed with the circInteractome algorithm [13] (see Supplemental Table 1 for primer sequences) in an Applied Biosystems by the qRT-PCR system. Fold-change analysis between sample groups was calculated by the Delta-Ct method.
Functional enrichment
Information about circRNAs is available on the circBase website (http://www.circbase.org/). The Circular RNA Interactome (https://circinteractome.nia.nih.gov/) was used to predict miRNAs and RBP-binding sites. The regulatory network was performed with Navigator software [14]. The set of RBPs common to all the differentially expressed circRNAs was analyzed with STRING: functional protein association networks (https://string-db.org) [15]. The set of miRNAs common to all the differentially expressed circRNAs was analyzed with miRNet 2.0 (https://www.mirnet.ca/miRNet/home.xhtml).
Statistical analysis
Continuous variables are expressed as the mean ± standard deviation. Categorical variables are expressed in frequency and percentage (%). Analysis of variance was applied to compare intergroup circRNAs levels. The Pearson correlation was used to test the link between echocardiographic and clinical variables vs. log2 circRNAs. In addition, the association between circRNAs and echocardiography parameters was assessed using logistic bivariate regression. Several models were constructed using the Wilcoxon test and iterating combinations between our circRNA candidates, as well as echocardiographic and clinical covariates. The changes in p-values of their variables were evaluated by the Wald test and a likelihood ratio. To characterize the diagnostic performance of the circRNAs candidate, ROC curves were applied together with a logistic regression model to determine the AUC and the specificity and sensitivity of the optimal cutoffs. ROC curves were generated by plotting sensitivity against 100-specificity. Data were presented as the AUC and 95% CI. The statistical software package R (www.r-project.org) was used for all analyses.
Results
Analysis of circRNA expression profiles in plasma of DCM patients
A total of 36 idiopathic and non-idiopathic DCM age-matched patients were assessed to test the differences in circRNA expression profiles (see Supplementary Figs. 1 and 2). A total of ten candidate circRNAs (see Supplementary Table 2) were obtained from circRNA microarray screening of plasmatic samples (fold change > 2, p < 0.05).
Validating the expression of the candidate circRNAs
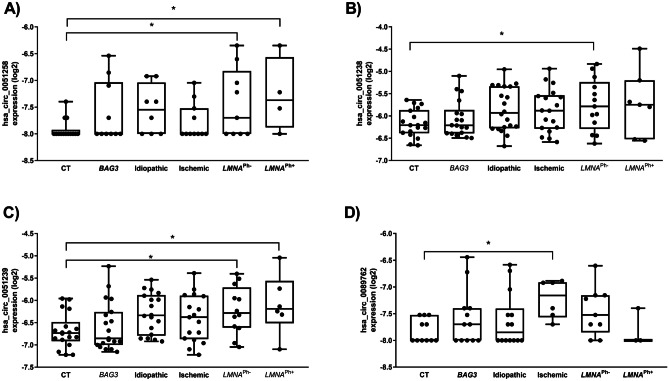
The expression of the ten circRNA candidates was carried out in plasma samples of each study group, using qRT-PCR. Only LMNA and ischemic DCM populations showed differential circRNA expression (Table 2). Circulating levels of hsa_circ_0051238, hsa_circ_0051239, and hsa_circ_0003258 were highly upregulated in the LMNA population compared to healthy controls (Fig. 2). To assess the strength of circRNAs as an early biomarker before the clinical manifestation of malignant ventricular arrhythmias and LV dilation, the LMNA-related DCM group was subdivided into LMNA pathogenic variant carrier, phenotypically negative (LMNAPh−) and phenotypically positive (LMNAPh+). Circulating hsa_circ_0003258 levels were differentially expressed in the LMNAPh− (LMNAPh− p = 0.03, LMNAPh+ p = 0.03) (Fig. 2A). The hsa_circ_0051238 levels were differentially expressed in the LMNAPh− population (p = 0.03) (Fig. 2B). And, the hsa_circ_0051239 plasmatic levels were significantly higher in both LMNA groups (LMNAPh− p = 0.03, LMNAPh+ p = 0.04) than in healthy subjects (Fig. 2C). Regarding the ischemic DCM cohort, the plasma hsa_circ_0089762 levels were significantly higher (p = 0.04) than in healthy subjects (Fig. 2D).
Table 2.
Peripheral circRNA levels in the study groups
| circRNA | CT | BAG3 DCM | Idiopathic DCM | Ischemic DCM | LMNAPh− DCM | LMNAPh+ DCM | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Med | Q1 | Q3 | Med | Q1 | Q3 | p | Med | Q1 | Q3 | p | Med | Q1 | Q3 | p | Med | Q1 | Q3 | p | Med | Q1 | Q3 | p | |
| hsa_circ_0003258 | − 8 | − 8 | − 7.92 | − 8 | − 8 | − 7.03 | 0.45 | − 7.55 | − 8 | − 7.04 | 0.29 | − 8 | − 8 | − 7.52 | 0.94 | − 7.69 | − 8 | − 6.82 | 0.03 | − 7.37 | − 7.88 | − 6.57 | 0.03 |
| hsa_circ_0051238 | − 6.20 | − 6.39 | − 5.9 | − 6.21 | − 6.39 | − 5.94 | 0.98 | − 5.90 | − 6.25 | − 5.33 | 0.10 | − 5.87 | − 6.30 | − 5.51 | 0.18 | − 5.71 | − 6.10 | − 5.18 | 0.08 | − 5.71 | − 5.98 | − 5.02 | 0.03 |
| hsa_circ_0051239 | − 6.73 | − 6.92 | − 6.48 | − 6.85 | − 7 | − 6.26 | 0.96 | − 6.33 | − 6.79 | − 5.88 | 0.09 | − 6.33 | − 6.87 | − 5.89 | 0.34 | − 6.28 | − 6.61 | − 5.71 | 0.03 | − 6.19 | − 6.51 | − 5.56 | 0.04 |
| hsa_circ_0089762 | − 8 | − 8 | − 7.52 | − 7.69 | − 8 | − 7.39 | 0.58 | − 7.84 | − 8 | − 7.40 | 0.75 | − 7.15 | − 7.57 | − 6.91 | 0.04 | − 7.52 | − 7.85 | − 7.15 | 0.25 | − 8 | − 8 | − 7.39 | > 0.9 |
Data presented as median (Q1–Q3). Coefficient significant at p < 0.05
BAG3 BCL2-associated athanogene 3, CT healthy control, DCM dilated cardiomyopathy, LMNA lamin A/C, LMNAPh− LMNA carrier of the pathogenic variant, LMNAPh+ LMNA carrier phenotypically positive, Med median
Fig. 2.
Boxplots of circRNA expression levels, normalized to MS2 RNA, in healthy subjects, BAG3-related DCM, idiopathic DCM, ischemic DCM, and LMNA-related DCM. The analysis was carried out using qRT-PCR. Data are present in log2. Data represent the mean ± SEM. *p < 0.05. Abbreviations: BAG3, BCL2-associated athanogene 3; CT, healthy cohort; circRNA, circular RNA; DCM, dilated cardiomyopathy; LMNA, lamin A/C; LMNAPh−, LMNA carrier of the pathogenic variant; LMNAPh+, LMNA carrier phenotype positive
Diagnostic value of the validated circRNAs in a DCM population
The receiver operating characteristic (ROC) area under the curve (AUC) analysis was assessed to investigate the circulating circRNAs diagnostic value in discriminating LMNA and ischemic DCM etiology from healthy controls. All individual circRNAs show an AUC ≥ 0.7. The highest AUC values reached by hsa_circ_0089762 that demonstrated an AUC value of 0.92 (95% of confidence intervals [CI] range of specificities are shown in Table 3).
Table 3.
Comparisons of single circRNA as predictors of DCM
| DCM etiology | circRNA | AUC (95% CI) | Sensitivity (%) | Specificity (%) | p |
|---|---|---|---|---|---|
| LMNA | hsa_circ_0003258 | 0.75 (0.56–0.94) | 61.53 | 78.57 | 0.043 |
| hsa_circ_0051238 | 0.71 (0.53–0.88) | 70 | 72.73 | 0.02 | |
| hsa_circ_0051239 | 0.73 (0.61–0.93) | 83.33 | 72.23 | 0.007 | |
| Ischemic | hsa_circ_0089762 | 0.92 (0.77–1) | 83.33 | 72.73 | 0.006 |
AUC area under the curve, CI confidence interval, DCM dilated cardiomyopathy, LMNA lamin A/C
Association between the expression of circRNAs and the clinical characteristics of the DCM population
The association between circulating circRNAs and echocardiographic and clinical features of DCM patients was also analyzed. As indicated in Table 4, the LMNAPh− group showed a negative correlation between hsa_circ_0003258 and hsa_circ_0051239 with early diastolic mitral annular velocity (E’s TDI). The LMNAPh+ cohort showed a positive correlation of hsa_circ_0051238 with tissue Doppler imaging (TDI) septal atrial systolic mitral annular velocity (A’s TDI) and a negative correlation of hsa_circ_0051238 and hsa_circ_0051239 with LV outflow tract (LVOT) velocity.
Table 4.
Correlation between the echocardiographic variables and individual circRNA for the LMNA cohort
| hsa_circ_0003258 | hsa_circ_0051238 | hsa_circ_0051239 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LMNAPh− | LMNAPh+ | LMNAPh− | LMNAPh+ | LMNAPh− | LMNAPh+ | |||||||||||||
| Pearson r | p | Power | Pearson r | p | Power | Pearson r | p | Power | Pearson r | p | Power | Pearson r | p | Power | Pearson r | p | Power | |
| A’s TDI (cm/s) | 0.191 | 0.623 | 0.677 | 0.676 | 0.324 | 0.584 | − 0.013 | 0.965 | 0.966 | 0.859 | 0.028 | 0.928 | 0.051 | 0.875 | 0.882 | 0.698 | 0.190 | 0.554 |
| E’s TDI (cm/s) | − 0.685 | 0.042 | 0.543 | − 0.413 | 0.587 | 0.7 | − 0.523 | 0.067 | 0.529 | − 0.157 | 0.766 | 0.793 | − 0.722 | 0.008 | 0.736 | − 0.253 | 0.681 | 0.265 |
| LVOT (cm/s) | − 0.085 | 0.856 | 0.87 | − 0.995 | 0.064 | 0.927 | − 0.020 | 0.963 | 0.8 | − 0.977 | 0.004 | 0.965 | 0.043 | 0.919 | 0.564 | − 0.97 | 0.030 | 0.923 |
A’s TDI atrial septal mitral annular velocity, DCM dilated cardiomyopathy, E’s TDI early diastolic mitral annular velocity, LMNA lamin A/C, LMNAPh− LMNA carrier of the pathogenic variant, LMNAPh+ LMNA carrier phenotypically positive, LVOT left ventricular outflow tract velocity, TDI tissue Doppler imaging
An additional study was performed to assess correlations between the echocardiographic and clinical variables and hsa_circ_0089762 for the ischemic DCM population. Hsa_circ_0089762 expression was negatively associated with diastolic blood pressure and LVEF (see Table 5).
Table 5.
Correlation between the clinical parameters and hsa_circ_0089762 for ischemic DCM cohort
| hsa_circ_0089762 | |||
|---|---|---|---|
| Pearson r | p | Power | |
| DBP (mm Hg) | -0.84 | 0.036 | 0.556 |
| LVEF (%) | -0.842 | 0.036 | 0.561 |
DCM dilated cardiomyopathy, DBP diastolic blood pressure, LVEF left ventricle ejection fraction
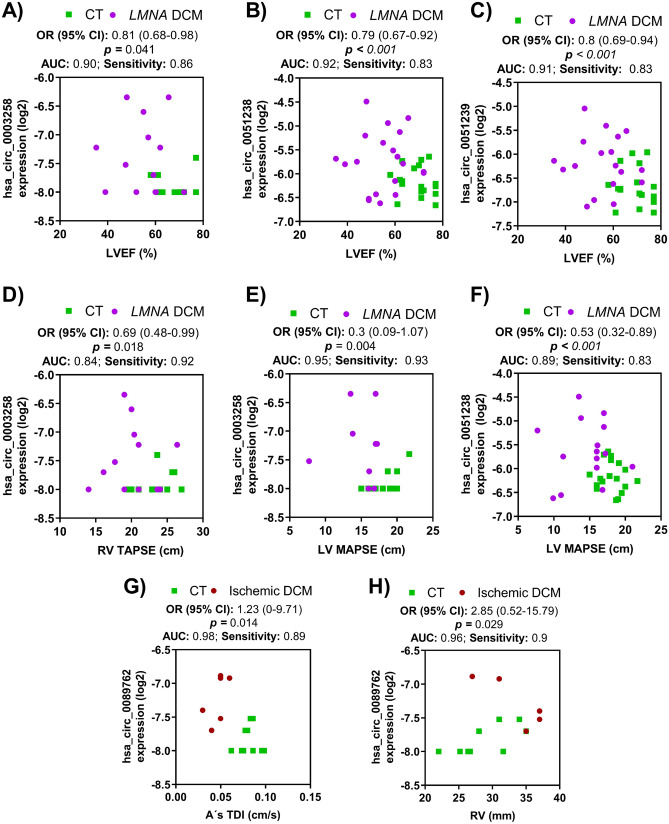
To further explore the expression of circRNA-DCM disease association, a logistic regression analysis was carried out in our DCM population (Fig. 3). All three LMNA-linked circRNAs were significantly related to male gender hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239. In the LMNA cohort, the bivariate logistic regression analyses revealed that all LVEF were independently negatively associated with hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239. LV mitral annular plane systolic excursion was independent negatively associated with hsa_circ_0003258 and hsa_circ_0051238. Right ventricle (RV) tricuspid annular plane systolic excursion was only independently negatively associated with hsa_circ_0003258. Pulmonary hypertension (PHT) was independently positively related to hsa_circ_0003258 and hsa_circ_0051239.
Fig. 3.
Bivariate logistic regression analysis for LMNA-related DCM and ischemic DCM patients. A–F Logistic regression analysis for the LMNA-related DCM cohort. LVEF was independently negatively related with hsa_circ_0003258 (A), hsa_circ_0051238 (B), and hsa_circ_0051239 (C). D RV tricuspid annular plane systolic excursion (TAPSE) was negatively related to hsa_circ_0003258. E, F LV mitral annular plane systolic excursion (MAPSE) was negatively correlated with hsa_circ_0003258 and hsa_circ_0051238. G, H The levels of hsa_circ_0089762 were associated with A’s TDI (G) and RV (H). The odds ratio, 95% of CI, and p values are indicated for each logistic regression analysis. Abbreviations: A’s TDI, atrial septal mitral annular velocity; AUC, area under the curve; CT, healthy group; CI, confidence intervals; LMNA, lamin A/C gene; LVEF, left ventricle ejection fraction; MAPSE, mitral annular plane systolic excursion; OR, odd ratio; RV; right ventricle; TAPSE, tricuspid annular plane systolic excursion
In the case of hsa_circ_0089762, the logistic regression analysis showed that its circulating levels within A’s TDI wave or the RV dimension were independent influencing factors for ischemic DCM.
Annotation for circRNA/RBPs interaction
An examination of biological processes related to RBPs, with binding sites for circRNA candidates, reveals a set of possible pathways in which circRNAs play a regulative role. LMNA mutation influences the proper development of megakaryocytes resulting in altered platelet production/function [16]. We recovered this LMNA effect in the enrichment (GO:0,045,652), regulation of megakaryocyte differentiation (FDR = 0.0013), and fibroblast growth (GO:0,008,543) (FDR = 0.0267).
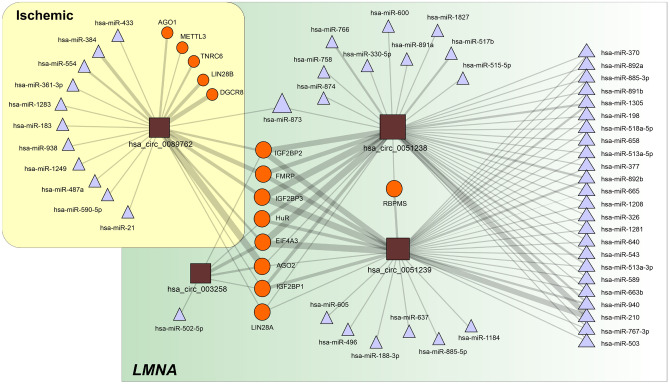
The analysis of the intersection set of RBPs predicted to interact with the selected circRNAs (Fig. 4; Table 6) shows clear enrichment in proteins involved in the control of transcriptional and translational processes (Table 7). Note the association with the regulation of membrane potential, in which IEF4A3 and FMRP are involved (FDR = 0.045).
Fig. 4.
CircRNA-centered regulatory network established among the selected circRNAs. The depicted interactions are based on data extracted from the circInteractome database and include miRNAs and RBPs. CircRNAs are represented as squares, RBPs as circles and miRNAs as triangles. The size of each symbol is proportional to the number of interactions established. The edge thickness is also proportional to the number of targets for each interacting partner as included in the circInteractome database. The regulatory network was prepared with Navigator software [14]. Abbreviations: DCM, dilated cardiomyopathy; LMNA, lamin A/C gene; miRNA, microRNA; RBP, RNA-binding protein
Table 6.
Differentially expressed circRNA potentially interact with RBP and miRNAs
| DCM etiology | circRNA | RBP | Predicted miRNAs target |
|---|---|---|---|
| LMNA | hsa_circ_003258 | AGO2, EIF4A3, HuR, IGF2BP1, IGF2BP2 | hsa-miR-502-5p |
| hsa_circ_0051238 | AGO2, EIF4A3, FMRP, HuR, IGF2BP1, IGF2BP2, IGF2BP3, LIN28A, RBPMS, ZC3H7B | hsa-miR-1208, hsa-miR-1281, hsa-miR-1305, hsa-miR-1827, hsa-miR-198, hsa-miR-210, hsa-miR-326, hsa-miR-330-5p, hsa-miR-370, hsa-miR-377, hsa-miR-503, hsa-miR-513a-3p, hsa-miR-513a-5p, hsa-miR-515-5p, hsa-miR-517b, hsa-miR-518a-5p, hsa-miR-543, hsa-miR-589, hsa-miR-600, hsa-miR-640, hsa-miR-658, hsa-miR-663b, hsa-miR-665, hsa-miR-758, hsa-miR-766, hsa-miR-767-3p, hsa-miR-873, hsa-miR-874, hsa-miR-885-3p, hsa-miR-891a, hsa-miR-891b, hsa-miR-892a, hsa-miR-892b, hsa-miR-940 | |
| hsa_circ_0051239 | AGO2, EIF4A3, FMRP, HuR, IGF2BP1, IGF2BP2, IGF2BP3, LIN28A, RBPMS | hsa-miR-1184, hsa-miR-1208, hsa-miR-1281, hsa-miR-1305, hsa-miR-188-3p, hsa-miR-198, hsa-miR-210, hsa-miR-326, hsa-miR-370, hsa-miR-377, hsa-miR-496, hsa-miR-503, hsa-miR-513a-3p, hsa-miR-513a-3p, hsa-miR-513a-5p, hsa-miR-518a-5p, hsa-miR-543, hsa-miR-589, hsa-miR-605, hsa-miR-637, hsa-miR-640, hsa-miR-658, hsa-miR-663b, hsa-miR-665, hsa-miR-767-3p, hsa-miR-885-3p, hsa-miR-885-5p, hsa-miR-891b, hsa-miR-892a, hsa-miR-892b, hsa-miR-940 | |
| Ischemic | hsa_circ_0089762 | AGO1, AGO2, DGCR8, EIF4A3, FMRP, IGF2BP1, IGF2BP2, IGF2BP3, LIN28A, LIN28B, METTL3, TNRC6 | hsa-miR-183, hsa-miR-21, hsa-miR-361-3p, hsa-miR-433, hsa-miR-590-5p |
DCM dilated cardiomyopathy, LMNA lamin A/C, RBP RNA-binding protein
Table 7.
Pathway analysis main findings: PPI enrichment analysis
| Pathway | Overlap | FDR | Genes | Database |
|---|---|---|---|---|
| Regulation of translation | 7/327 | 2.09e − 10 | AGO2,EIF4A3,ELAVL1,FMRP,IGF2BP1,IGF2BP2,IGF2BP3 | GO |
| mRNA binding | 7/198 | 1.31e − 12 | AGO2,EIF4A3,ELAVL1,FMRP,IGF2BP1,IGF2BP2,IGF2BP3 | GO |
| Negative regulation of nitrogen compound metabolic process | 7/2307 | 8.81e − 06 | AGO2,EIF4A3,ELAVL1,FMRP,IGF2BP1,IGF2BP2,IGF2BP3 | GO |
| Regulation of mRNA stability | 5/113 | 1.05e − 08 | ELAVL1,FMRP,IGF2BP1,IGF2BP2,IGF2BP3 | GO |
| mRNA transport | 5/148 | 3.12e − 08 | EIF4A3,FMRP,IGF2BP1,IGF2BP2,IGF2BP3 | GO |
| Regulation of gene silencing by miRNA | 3/78 | 3.82e − 05 | AGO2,ELAVL1,FMRP | GO |
| Regulation of membrane potential | 2/408 | 0.0445 | EIF4A3,FMRP | GO |
| MAPK6/MAPK4 signaling | 2/86 | 0.0077 | AGO2,IGF2BP1 | GO |
| ncRNA processing | 2/340 | 0.0344 | AGO2,EIF4A3 | GO |
PPI enrichment p-value:1.67e − 10
FDR false discovery rate, GO gene ontology, KW keyword
The analysis of miRNAs sponged by validated circRNAs offers various candidates for further research. Hsa_circ_0003258 has only one functional binding site to hsa-miR-653. As a counterpart, hsa_circ_0051238 and hsa_circ_0051239 present a clear sponge effect over hsa-miR-210, with five binding sites that have ∆U below zero. Thereby, the overexpression of hsa_circ_0051238 and hsa_circ_0051239 will actively reduce the availability of hsa-miR-210. Hsa-miR-210 regulates expression of hepatocyte growth factor gene, whose overexpression is considered a treatment for DCM [17]. Additionally, they also present a functional binding site for hsa-miR-330-5p that is involved in cardiomyocyte survival and function recovery [18]. Regarding miRNA-related diseases, hsa_circ_0051238 sponges hsa-miR-873 and hsa-miR-513a-5p are both related with heart disease (p = 0.075), and hsa-miR-377 is related with ischemic cardiomyopathy (p = 0.221). Hsa_circ_0089762 has sponge activity with multiple, energetically favorable binding sites. Of note is hsa-miR-21, as well as hsa-miR-183, hsa-miR-361-3p, hsa-miR-384, hsa-miR-873, hsa-miR-938, hsa-miR-1249, and hsa-miR-1283. The miRNAs sponged by the circRNAs with a context score over 90% was used to capture the set of mRNAs regulated by these miRNAs. Functional enrichment, using a hypergeometric association algorithm, shows that 148 proteins of the network were related with focal adhesion (p = 2.68e−8), and 128 proteins were linked with regulation of the actin cytoskeleton (p = 0.00002). Gene ontology biological processes, using the same hypergeometric algorithm, show a significant correlation with endoplasmic reticulum-nuclei signaling pathways (p = 0.1e−6) and pre- and post-Golgi vesicle transportation (p = 8.6e−7 and 4.47e−7, respectively).
Discussion
Over the last decade, the diagnostic process of DCM etiologies has focused on searching for new biomarkers. An efficient biomarker for DCM should be robust, stable, non-invasive, sensitive, specific to this entity, predictive of a particular DCM etiology, and show a preclinical and clinical relevance to be validated in animal and/or human cell models [19]. We propose the use of peripheral circRNAs as a novel discriminant biomarker of DCM etiologies.
Unlike linear RNA, single circulating circRNAs or circRNAs combined with various other biomarkers are a promising tool for clinical diagnosis of heart diseases, which would improve outcome [20]. Thus, circRNA MICRA was reported to risk-stratify patients after acute myocardial infarction [21]. Peripheral circ_0124644 and circ_0098964 levels have been described as a diagnostic biomarker of coronary artery disease [22]. Related to cardiomyopathies, a set of circulating circRNAs DNAJC6, TMEM56, and MBOAT2 has been proposed to discriminate between healthy and hypertrophic cardiomyopathy [23]. In this sense, hsa_circ_0071542 was upregulated in children with fulminant myocarditis in leukocytes isolated from peripheral blood [24]. Nevertheless, this area remains mostly unexplored in DCM [22, 25]. Recent studies have described several circRNA expression profiles in the DCM population compared to healthy patients. However, to date, it has not been studied among the different etiologies of DCM [26, 27]. Hence, further analysis of circRNAs among DCM etiologies might provide early, precise characterization of the disease and lead to novel pathological information, beyond the traditional biomarkers. To the best of our knowledge, the present study is the first to describe a subset of circulating circRNA for a discriminative etiology-based diagnostic in DCM. Circulating hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239 expression levels were upregulated in LMNA-related DCM patients. Notably, hsa_circ_0051238 plasmatic levels were significantly present in the LMNAPh− cohort. Hence, it may be a promising diagnostic biomarker for the early identification of patients in an initial stage of LMNA-related DCM. This will allow personalized therapeutic measures to be applied that help to improve the progression and outcome of LMNA-related DCM. Furthermore, plasmatic hsa_circ_0089762 may provide discriminative power for the ischemic DCM cohort with high-yield diagnostic accuracy and an AUC of 0.92. These circRNAs have been identified mostly in various types of oncologic processes [28–33]. Thus, only hsa_circ_0051239 levels have been upregulated in the myocardium of congenital ventricular septal defect [33]. However, they have not been previously described in DCM cases.
In the current study, circRNA were related to clinical and echocardiographic variables. Male gender, rare non-missense variants in LMNA and LVEF < 50% have been established as independent factors associated with a more aggressive outcome and even death during follow-up [34]. Herein, all three circRNAs associated with LMNA-DCM etiology were related to male gender [35]. On the other side, echocardiography variables and related circRNAs might suggest a time-evolving sequence. TDI echocardiography is a non-invasive, very sensitive method to assess the cardiac hemodynamic in DCM [36]. TDI reveals that subtle impairments in diastolic myocardial tissue velocities are markers of early cardiac disease and have been associated with outcome in various cardiopathies [16, 17]. In the LMNAPh− group, the E’s TDI is negatively related to hsa_circ_0003258 and hsa_circ_0051239. This E’s TDI impairment suggests an underlying early diastolic dysfunction [37]. A’s TDI in the LMNAPh+ group showed a positive correlation, which indicates that the left atrium is a prominent factor to maintain the LV filling pressure when diastolic dysfunction advances. This sequential TDI septal impairment mirrors the transition from LMNAPh− to LMNAPh+ and may be related to the progressive fibrosis of the interventricular septum located in the basal portion, which is characteristic of the LMNA related-DCM that has been associated with ventricular arrhythmias and worse prognosis [38]. LVEF was independently negatively associated with hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239. According to the LV systolic impairment, hsa_circ_0003258 and hsa_circ_0051238 were related to LV mitral annular plane systolic excursion. Thus, changes in contractility quantified by LV mitral annular plane systolic excursion occur as compensatory mechanisms before impairment of ventricular function [39]. Hsa_circ_0051238 and hsa_circ_0051239 were also negatively related to LVOT velocity, which suggests progressive impairment of the cardiac pump in the LMNAPh+ cohort. Dysfunction of RV is a final common step in DCM and heart failure [40]. RV pressure overload due to PHT and the interventricular interdependence affected by septal fibrosis and underlying ischemia may influence this situation. In support of our results, circRNA, hsa_circ_0003258 was positively increased with the RV lower tricuspid annular plane systolic excursion and PHT [41].
Otherwise, hsa_circ_0089762 correlated to diastolic blood pressure and LVEF in the ischemic group, which supports our results as a specific, highly sensitive biomarker with high-yield diagnostic accuracy. Moreover, hsa_circ_0089762 was related to A’s TDI, which suggests more advanced progression of this entity. Its association with an increase in RV dimension could add information for tailored management in this group, since RV impairment is a worse outcome marker in the ischemic population [42]. In addition, RV involvement has a multifactorial origin that may be influenced by LV remodelling, increased LV filling pressures, and the appearance of PHT or RV ischemia [43].
Regarding biological implications, circRNAs spring from introns or exons of their parental genes by back-spliced circularization [25]. Hence, the ratio between linear and circular fractions affects gene expression. According to the protein atlas (proteinatlas.org), parental genes are expressed in cardiac tissue, which supports correlations between etiologies and circRNAs. Hsa_circ_0003258 is synthesized from ZNF652 gene. ZNF652 interacts with CBFA2T3, which acts as a transcriptional repressor [44]. ZNF652 is associated with systolic or diastolic blood pressure and hypertension. However, its role remains unclear [45]. Hsa_circ_0051238 and hsa_circ_0051239 come from the ATP5SL gene. ATP5SL is required for the assembly of mitochondrial NADH: ubiquinone oxidoreductase complex (complex I). Complex I is essential to provide the energy for cardiac function and is related to DCM progression [46]. ATP5SL has been associated with a congenital ventricular septal defect by the overexpression of hsa_circ_0051239 [47]. Finally, hsa_circ_0089762 is generated from the MT-CO2 gene. MT-CO2 is part of the electron transport chain of the mitochondria. Reduced activity of the electron transport chain subunits has been described independently of etiology in ischemic or idiopathic DCM patients [48].
The functional enrichment of the intersecting set of RBSs reveals the role of FMRP in regulation of the membrane potential. Bao et al. described FMRP isoform 1, in rats, as an essential protection factor and a novel potential biomarker in the cardiovascular system [49]. The participation of circRNAs in regulatory networks involving competing-endogenous RNA interactions by sequestering miRNAs has been characterized recently in cardiovascular pathologies [50, 51]. From the set of miRNAs that could be sponged by the circRNAs that we considered, we found significant enrichment in the regulation of focal adhesion and actin cytoskeleton. Both have an important role in human DCM [52], which suggests new pathways of study.
Our current study has several limitations. Firstly, our sample was prospectively recruited from the outpatient clinic. The size of the study sample, comprised of strictly DCM patients, did not allow us to obtain a robust multivariate logistic regression model. Furthermore, a larger sample size is needed to validate these data by gender categorization since gender may play a role in the DCM prognosis [53, 54]. In consequence, these results should be extended and replicated to a larger population before the novel biomarkers can be routinely applied in clinical practice. Furthermore, data on natriuretic peptides or troponin were not accessible for all patients. Finally, even though databases registered the expression of the parental genes in cardiac tissue, we have no confirmation about the direct secretion from the heart of these circulating circRNAs into the extracellular space. Hence, the association of circRNAs with DCM and all the interactions are putative. Further analysis should be carried out on human heart samples to confirm our results.
Conclusion
Exploring new biomarkers through circular transcriptome expression patterns will identify new targets in DCM pathogenesis. We propose a circulating circRNAs fingerprint to discriminate between various DCM etiologies. Circulating hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239 expression levels are higher in LMNA-related DCM, and hsa_circ_0089762 levels are specifically upregulated in the ischemic DCM cohort. These circulating circRNAs and certain echocardiographic variables might improve the etiology-based diagnostic, which allows early identification of asymptomatic cases and tailored treatment of the DCM population.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Galan Pacheco for statistical support. We would also like to thank all patients involved in this project.
Abbreviations
- A’s TDI
Atrial septal mitral annular velocity
- AUC
Area under the curve
- BAG3
BCL2-associated athanogene 3
- CI
Confidence interval
- circRNA
Circular RNA
- DCM
Dilated cardiomyopathy
- E’s TDI
Early diastolic mitral annular velocity
- FDR
False discovery rate
- LMNA
Lamin A/C
- LV
Left ventricular
- LVEF
LV ejection fraction
- LVOT
LV outflow tract
- MAPSE
Mitral annular plane systolic excursion
- mRNA
Messenger RNA
- miRNA
MicroRNA
- PHT
Pulmonary hypertension
- ROC
Receiver operating characteristic
- RBP
RNA binding protein
- qRT-PCR
Quantitative real-time polymerase chain reaction
- TAPSE
Tricuspid annular plane systolic excursion
- TDI
Tissue Doppler imaging
Author contribution
All authors have read and approved the submission of the manuscript. Calderon-Dominguez M, Mangas A, and Toro R conceived the experiments; Quezada-Feijoo M, Ramos M, Campuzano O, Sarquellas Brugada-G, Pinilla JM, Robles Mezcua A, Mangas A, and Toro R recruited the subjects; and Thalia extracted RNA from samples. Costa M, Calderon-Dominguez M, Pacheco-Cruz GA, Enguita FJ, and Toro R conducted the experiments and analyzed the results. Calderon-Dominguez M, Mangas A, and Toro R wrote the manuscript. All authors reviewed the manuscript.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by grants in the framework of the European Regional Development Fund (ERDF) Integrated Territorial Initiative (ITI PI0048-2017 and ITI0033_2019), a clinical research grant from the Spanish Society of Cardiology for Basic Research in cardiology (PI0012_2019), COST (European Cooperation in Science and Technology) Action EUCardioRNA CA17129, and the Portuguese Foundation for Science and Technology (FCT) under the framework of the research grant PTDC-MED-GEN-29389–2017.
Data availability
Data transparency is guaranteed. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
We used various softwares for functional enrichment and statistical analysis. All of them are cited in our manuscript.
Declarations
Conflict of interests
We know of no conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome. The study protocol was approved by the Andalusian Biomedical Research Ethics committee. The study was performed in full compliance with the Declaration of Helsinki. Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients for paper publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Calderon-Dominguez, Email: mariacalderond@gmail.com.
Alipio Mangas, Email: alipio.mangas@uca.es.
Oscar Campuzano, Email: oscar@brugada.org.
Georgia Sarquella-Brugada, Email: georgia@brugada.org.
Mónica Ramos, Email: monica.ramos81@gmail.com.
Maribel Quezada-Feijoo, Email: maribelquezada2000@gmail.com.
Ainhoa Robles-Mezcua, Email: ainhoa.mezcua@gmail.com.
Galan del Aguila Pacheco-Cruz, Email: marlucale41@gmail.com.
Thalia Belmonte, Email: thaliabelmonte@gmail.com.
Francisco J. Enguita, Email: fenguita@medicina.ulisboa.pt
Rocío Toro, Email: rociotorogreen@gmail.com.
References
- 1.Savarese G, Lund LH (2017) Global public health burden of heart failure. Card Fail Rev 03:7. 10.15420/cfr.2016:25:2 [DOI] [PMC free article] [PubMed]
- 2.McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbaum AN, Agre KE, Pereira NL. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020;17:286–297. doi: 10.1038/s41569-019-0284-0. [DOI] [PubMed] [Google Scholar]
- 4.Van Der Bijl P, Delgado V, Bootsma M, Bax JJ. Risk stratification of genetic, dilated cardiomyopathies associated with neuromuscular disorders. Circulation. 2018;137:2514–2527. doi: 10.1161/CIRCULATIONAHA.117.031110. [DOI] [PubMed] [Google Scholar]
- 5.Domínguez F, Cuenca S, Bilińska Z, et al. Dilated cardiomyopathy due to BLC2- associated athanogene 3 (BAG3) mutations. J Am Coll Cardiol. 2018;72:2471–2481. doi: 10.1016/j.jacc.2018.08.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S, Baldinger SH, Gandjbakhch E, et al. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. J Am Coll Cardiol. 2016;68:2299–2307. doi: 10.1016/j.jacc.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 7.Lu D, Thum T. RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat Rev Cardiol. 2019;16:661–674. doi: 10.1038/s41569-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stepien E, Costa MC, Kurc S, et al. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases review-article. Acta Pharmacol Sin. 2018;39:1085–1099. doi: 10.1038/aps.2018.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies: a position statement from the european society of cardiology working group on myocardial and pericardial diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 11.Belmonte T, Mangas A, Calderon-Dominguez M, et al. Peripheral microRNA panels to guide the diagnosis of familial cardiomyopathy. Transl Res. 2020;218:1–15. doi: 10.1016/j.trsl.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Toro R, Blasco-Turrión S, Morales-Ponce FJ, et al. Plasma microRNAs as biomarkers for lamin A/C-related dilated cardiomyopathy. J Mol Med. 2018;96:845–856. doi: 10.1007/s00109-018-1666-1. [DOI] [PubMed] [Google Scholar]
- 13.Dudekula DB, Panda AC, Grammatikakis I, et al. Circinteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome – using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE. 2011;6:e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izquierdo I, Rosa I, Bravo SB, et al. Proteomic identification of putative biomarkers for early detection of sudden cardiac death in a family with a LMNA gene mutation causing dilated cardiomyopathy. J Proteomics. 2016;148:75–84. doi: 10.1016/j.jprot.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Komamura K, Tatsumi R, Miyazaki J et al (2004) Treatment of dilated cardiomyopathy with electroporation of hepatocyte growth factor gene into skeletal muscle. Hypertens (Dallas, Tex 1979) 44:365–71. 10.1161/01.HYP.0000139916.96375.47 [DOI] [PubMed]
- 18.Van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martín-Ventura JL, Blanco-Colio LM, Tuñón J et al (2009) Biomarkers in cardiovascular medicine. Rev Española Cardiol (English Ed 62:677–688. 10.1016/s1885-5857(09)72232-7 [DOI] [PubMed]
- 20.Khan MAF, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 21.Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. IJC Hear Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa-circ-0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:1–9. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnenschein K, Wilczek AL, de Gonzalo-Calvo D, et al. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci Rep. 2019;9:1–8. doi: 10.1038/s41598-019-56617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Han B, Wang J, et al. Differential expression profiles and functional analysis of circular RNAs in children with fulminant myocarditis. Epigenomics. 2019;11:1129–1141. doi: 10.2217/epi-2019-0101. [DOI] [PubMed] [Google Scholar]
- 25.Gabriel AF, Costa MC Enguita FJ (2020) Circular RNA-centered regulatory networks in the physiopathology of cardiovascular diseases. Int J Mol Sci 21 [DOI] [PMC free article] [PubMed]
- 26.Sun W, Han B, Cai D, et al. Differential expression profiles and functional prediction of circular RNAs in pediatric dilated cardiomyopathy. Front Mol Biosci. 2020;7:600170. doi: 10.3389/fmolb.2020.600170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Z, Zhao Y, Dai F, et al. Analysis of changes in circular RNA expression and construction of ceRNA networks in human dilated cardiomyopathy. J Cell Mol Med. 2021;25:2572–2583. doi: 10.1111/jcmm.16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Duan H, Li Y, et al. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun. 2019;513:812–819. doi: 10.1016/j.bbrc.2019.03.214. [DOI] [PubMed] [Google Scholar]
- 29.Fu C, Lv R, Xu G, et al. Circular RNA profile of infantile hemangioma by microarray analysis. PLoS ONE. 2017;12:1–16. doi: 10.1371/journal.pone.0187581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WL Mo JT Jiang L Zhang et al 2020 Circular RNA hsa_circ_0000467 promotes the development of gastric cancer by competitively binding to microRNA miR-326-3p. Biomed Res Int 2020 10.1155/2020/4030826 [DOI] [PMC free article] [PubMed]
- 31.Wu Z, Sun H, Wang C, et al. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol Ther - Nucleic Acids. 2020;20:801–811. doi: 10.1016/j.omtn.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yavropoulou M, Poulios C, Michalopoulos N, et al. A role for circular non-coding RNAs in the pathogenesis of sporadic parathyroid adenomas and the impact of gender-specific epigenetic regulation. Cells. 2018;8:15. doi: 10.3390/cells8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C, Yu Y, Ding F. Microarray analysis of circular RNA expression profiles associated with gemcitabine resistance in pancreatic cancer cells. Oncol Rep. 2018;40:395–404. doi: 10.3892/or.2018.6450. [DOI] [PubMed] [Google Scholar]
- 34.Van Rijsingen IAW, Arbustini E, Elliott PM, et al. Risk factors for malignant ventricular arrhythmias in lamin A/C mutation carriers: a European cohort study. J Am Coll Cardiol. 2012;59:493–500. doi: 10.1016/j.jacc.2011.08.078. [DOI] [PubMed] [Google Scholar]
- 35.van Rijsingen IAW, Nannenberg EA, Arbustini E, et al. Gender-specific differences in major cardiac events and mortality in lamin A/C mutation carriers. Eur J Heart Fail. 2013;15:376–384. doi: 10.1093/eurjhf/hfs191. [DOI] [PubMed] [Google Scholar]
- 36.Van Rijsingen IAW, Bakker A, Azim D, et al. Lamin A/C mutation is independently associated with an increased risk of arterial and venous thromboembolic complications. Int J Cardiol. 2013;168:472–477. doi: 10.1016/j.ijcard.2012.09.118. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Serra A, Toro R, Campuzano O, et al. A novel mutation in lamin A/C causing familial dilated cardiomyopathy associated with sudden cardiac death. J Card Fail. 2015;21:217–225. doi: 10.1016/j.cardfail.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Fontana M, Barison A, Botto N, et al. CMR-verified interstitial myocardial fibrosis as a marker of subclinical cardiac involvement in LMNA mutation carriers. JACC Cardiovasc Imaging. 2013;6:124–126. doi: 10.1016/j.jcmg.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez-Suarez DF, Lopez-Menendez F, Roche-Lima A, Lopez-Candales A (2019) Assessment of mitral annular plane systolic excursion in patients with left ventricular diastolic dysfunction. Cardiol Res 10:83–88. 10.14740/cr837 [DOI] [PMC free article] [PubMed]
- 40.Rudski LG, Lai WW, Afilalo J, et al. Guidelines for the echocardiographic assessment of the right heart in adults: a report from the American Society of Echocardiography. Endorsed by the European Association of Echocardiography, a registered branch of the European Society of Cardiology, and. J Am Soc Echocardiogr. 2010;23:685–713. doi: 10.1016/j.echo.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Merlo M, Gobbo M, Stolfo D, et al. The prognostic impact of the evolution of RV function in idiopathic DCM. JACC Cardiovasc Imaging. 2016;9:1034–1042. doi: 10.1016/j.jcmg.2016.01.027. [DOI] [PubMed] [Google Scholar]
- 42.Brieke A, DeNofrio D. Right ventricular dysfunction in chronic dilated cardiomyopathy and heart failure. Coron Artery Dis. 2005;16:5–11. doi: 10.1097/00019501-200502000-00002. [DOI] [PubMed] [Google Scholar]
- 43.Kukulski T, She L, Racine N, et al. Implication of right ventricular dysfunction on long-term outcome in patients with ischemic cardiomyopathy undergoing coronary artery bypass grafting with or without surgical ventricular reconstruction. J Thorac Cardiovasc Surg. 2015;149:1312–1321. doi: 10.1016/j.jtcvs.2014.09.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar R, Selth LA, Schulz RB, et al. Genome-wide mapping of ZNF652 promoter binding sites in breast cancer cells. J Cell Biochem. 2011;112:2742–2747. doi: 10.1002/jcb.23214. [DOI] [PubMed] [Google Scholar]
- 45.Korkor MT, Meng FB, Xing SY, et al. Microarray analysis of differential gene expression profile in peripheral blood cells of patients with human essential hypertension. Int J Med Sci. 2011;8:168–179. doi: 10.7150/ijms.8.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarreta D, Orús J, Barrientos A, et al. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc Res. 2000;45:860–865. doi: 10.1016/S0008-6363(99)00388-0. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Hu Y, Zhuang B, et al. Differential expression of CircRNAs in embryonic heart tissue associated with ventricular septal defect. Int J Med Sci. 2018;15:703–712. doi: 10.7150/ijms.21660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Govindaraj P, Rani B, Sundaravadivel P, et al. Mitochondrial genome variations in idiopathic dilated cardiomyopathy. Mitochondrion. 2019;48:51–59. doi: 10.1016/j.mito.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Bao J, Ye C, Zheng Z, Zhou Z. Fmr1 protects cardiomyocytes against lipopolysaccharide-induced myocardial injury. Exp Ther Med. 2018;16:1825–1833. doi: 10.3892/etm.2018.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Gan TY, Li N, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa MC, Cortez-Dias N, Gabriel A, et al. circRNA-miRNA cross-talk in the transition from paroxysmal to permanent atrial fibrillation. Int J Cardiol. 2019;290:134–137. doi: 10.1016/j.ijcard.2019.04.072. [DOI] [PubMed] [Google Scholar]
- 52.Towbin JA. The role of cytoskeletal proteins in cardiomyopathies. Curr Opin Cell Biol. 1998;10:131–139. doi: 10.1016/S0955-0674(98)80096-3. [DOI] [PubMed] [Google Scholar]
- 53.Yogasundaram H, Qi A, Nguyen Q, Oudit GY. Battle of the sexes: differential prognosis by sex in dilated cardiomyopathy. Can J Cardiol. 2020;36:7–10. doi: 10.1016/j.cjca.2019.06.031. [DOI] [PubMed] [Google Scholar]
- 54.Cannatà A, Fabris E, Merlo M, et al. Sex differences in the long-term prognosis of dilated cardiomyopathy. Can J Cardiol. 2020;36:37–44. doi: 10.1016/j.cjca.2019.05.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data transparency is guaranteed. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
We used various softwares for functional enrichment and statistical analysis. All of them are cited in our manuscript.