Abstract
Acanthamoeba spp. are among the most worldwide prevalent protozoa. It is the causative agent of a disease known as Acanthamoeba keratitis, a painful and severe sight-threatening corneal infection that can lead to blindness. In recent years, the prevalence of Acanthamoeba keratitis has rapidly increased, growing its importance to human health. This systematic review aims to assess the distribution of Acanthamoeba sp. genotypes causing keratitis around the world, considering the sample collected type and the used identification method. Most of the cases were found in Asia and Europe. Not surprisingly, the T4 genotype was the most prevalent worldwide, followed by T3, T15, T11, and T5. Furthermore, the T4 genotype contains a higher number of species. Given the differences in pathology, susceptibility to treatment, and clinical outcome between distinct genotypes, it is essential to genotype isolates from Acanthamoeba keratitis cases to help to establish a better correlation between in vitro and in vivo activities, resulting in better drug therapies and successful treatment in cases of this important ocular infection.
Keywords: Protozoan, Free-living amoebae, Acanthamoeba spp., Keratitis, Genotype
Introduction
Free-living amoebae (FLA) from the genus Acanthamoeba are ubiquitously distributed in nature (Siddiqui and Khan 2012). This parasite can be isolated from virtually any natural or artificial environment including soil, dust, water, air, medical equipment, lens fluids, air-conditioning, and nasopharyngeal mucosa from healthy individuals (Clarke and Niederkorn 2006; Nagyová et al. 2010a; Siddiqui and Khan 2012; Khezri et al. 2016; Tawfeek et al. 2016; Król-Turmińska and Olender 2017; Lass et al. 2017; Wopereis et al. 2020). Acanthamoeba spp. are among the most prevalent protozoa worldwide and are considered amphizoic organisms due to their ability to live either as a free-living organism or as pathogenic and opportunistic protozoa. This behavior enables Acanthamoeba sp. to live in close contact with potential hosts, including humans, leading to severe infections (Oddo 2006; Visvesvara et al. 2007; Lanocha et al. 2009).
Furthermore, these protozoa can disseminate many different pathogens besides causing infections by themselves. Acanthamoeba sp. protozoa are known as “Trojan horses,”' acting as a host for a wide range of microorganisms including viruses, protists, and bacteria, and in that way, being a reservoir for maintaining and dispersing those endosymbionts in the environment (Greub and Raoult 2004; Berger et al. 2006; Siddiqui and Khan 2012).
When Acanthamoeba sp. defeats the host barriers and establishes infection, it can result in a clinical condition called granulomatous amebic encephalitis (GAE). GAE is an opportunistic, insidious, and chronic infection of the central nervous system, which presents high mortality despite low incidence (Visvesvara et al. 2007; Visvesvara and Schuster 2008; Diaz 2010). These FLA can also cause several highly destructive and disseminating infections concerning lungs, kidneys, liver, adrenal glands, heart, bones, and skin, affecting both immunocompromised and immunocompetent patients. However, the most common extracerebral infection caused by this amoeba is Acanthamoeba keratitis (AK) (Khan 2006; Ren and Wu 2010; Walochnik et al. 2015; Kot et al. 2018). Acanthamoeba keratitis is a painful and severe sight-threatening corneal disease that can even lead to blindness. Unlike GAE, AK can also occur in immunocompetent individuals due to poor lens care or following a corneal trauma (Visvesvara et al. 2007; Dart et al. 2009).
The symptoms and clinical findings of AK include severe pain, considerable lacrimation, photophobia, inflammation, corneal abrasion and opacification, blurred vision, foreign body sensation, edema, stromal infiltration, epithelial loss, ring ulcers, cataract, glaucoma, and even corneal perforation and vision loss if not adequately treated (Khan 2006; Castrillón and Orozco 2013; Lorenzo-Morales et al. 2015). The same symptoms can occur in bacterial, fungal, and viral keratitis, commonly leading to a misdiagnosis. AK usually progresses slower than those infections (Lorenzo-Morales et al. 2013). Furthermore, coinfections of Acanthamoeba spp. with fungi as Fusarium and Candida, or bacteria as Pseudomonas have already been reported (Sharma et al. 2013; Nunes et al. 2016; Buchele et al. 2018).
In recent years, the prevalence of AK has rapidly increased due to the rising recognition of this infection as an essential threat to human health. This swelling in the prevalence, in part, is due to the availability of modern diagnostic methods that allow differential diagnosis from other keratitis, added to the increasing number of contact lens (CL) users, which is the main risk factor for AK (Khan 2006; Patel and Hammersmith 2008; Dart et al. 2009).
The number of CL users grows worldwide every year, and the number of AK cases is increasing concomitantly (Maycock and Jayaswal 2016). About 90% of patients diagnosed with AK are CL wearers, with reported rates between 1 and 33 cases per million (Khan 2006; Visvesvara et al. 2007). Considering that amoeba can reach lens cases through the air or tap water, AK infections are often related to poor cleaning, overuse, swimming, or sleeping wearing CL (Shoff et al. 2008; Walochnik et al. 2015). However, it is important to note that even patients who regularly disinfect their lens with a multipurpose solution can still contract AK since it has been shown that some commercially available cleaning solutions are ineffective against the protozoan (Kilvington et al. 2004; Hammersmith 2006; Shoff et al. 2008; Walochnik et al. 2015). One explanation for this phenomenon is the biofilm formation following lens contamination. The formed biofilm can enhance Acanthamoeba lifetime on lens storage added to provide nutrients for the amoeba, playing an essential role in the pathology of AK (Khan 2006). On top of this, there are more predisposing factors for AK, even for non-users of lenses, which include previous mechanical corneal trauma associated with exposure to contaminated soil, water, or vegetation (Jiang et al. 2006; Wesolowska et al. 2006; Lorenzo-Morales et al. 2015).
AK is one of the most challenging corneal diseases to be diagnosed. It is often only considered after the failure of the first-line therapy for herpes simplex virus or bacterial/fungal keratitis. In addition, currently diagnosis methods are invasive, requiring stromal biopsy or corneal scrapes, for example. Moreover, the sooner the disease is diagnosed, the better is the prognosis (Dart et al. 2009; Page and Mathers 2013).
AK treatment is difficult and long-termed, becoming an extremely challenging problem since there are no approved drugs for this infection specifically. Indeed, multiple antibacterial, antifungal, and antiamoebic agents are used in combination to improve the outcome (Gokhale 2008; Wilhelmus et al. 2008; Juárez et al. 2018). The challenges in treatment happen due to different factors, including the wide range of virulence showed by Acanthamoeba spp. which makes it challenging to establish a correlation between in vivo and in vitro drug activity. Furthermore, the cystic forms of Acanthamoeba sp. are extremely resistant. Even after a clinical cure, the treatment must continue for a long period to prevent relapses of the infection resulting from the sporulation of the remaining cysts (Kumar and Lloyd 2002; Astorga et al. 2011). When oral or topical treatments have failed in severe infections, corneal transplantation is the last therapeutic option (Kitzmann et al. 2009; Nguyen et al. 2010; Lorenzo-Morales et al. 2015). Among the measures that can be used to prevent the infection, one of the most important is to educate lens users regarding the proper care of contact lenses and its cases, the importance of appropriate disinfecting solutions, no overnight use, and no showering or swimming wearing CL, in order to avoid the contact with contaminated water (Visvesvara et al. 2007).
The life cycle of Acanthamoeba spp. consists of two stages: an actively feeding and reproductive trophozoite, and a latent cyst stage with minimal metabolic activity (Siddiqui and Khan 2012; Lorenzo-Morales et al. 2015). Trophozoites of Acanthamoeba spp. exhibit prominent vacuoles and typical acanthopodia, which are fine and spine-like structures on their surface. Their size usually is around 12–35 µm in diameter, but it can vary significantly between isolates due to the many different genotypes and species (Khan 2006; Visvesvara et al. 2007; Costa et al. 2010). The encystment occurs due to unfavorable environmental conditions such as desiccation, changes in pH and temperature, increased osmolarity or hypo-osmolarity, and food deprivation. The trophozoite becomes a metabolically inactive cyst with a double wall, composed of an endocyst and ectocyst, both containing cellulose (Marciano-Cabral and Cabral 2003; Munguía 2005; Siddiqui et al. 2012; Costa et al. 2010; Martín-Pérez et al. 2017). The cyst form allows the organism to survive to extreme conditions, retaining its pathogenic properties for long periods in hostile environments. Altogether, that explains why AK treatment is difficult because Acanthamoeba encysts when the environment becomes unfavorable due to the medications. Both cysts and trophozoites can adhere to the surface, including soft or rigid CL and contact lens cases, allowing the parasite to invade the eye tissue (Khan and Tareen 2003; Marciano-Cabral and Cabral 2003; Siddiqui and Khan 2012).
In an attempt to organize the increasing number of isolates belonging to the Acanthamoeba genus, Pussard and Pons (1977) initially classified the species based on morphological features of the cysts. Then, according to the ectocyst and endocyst shape and size, Acanthamoeba spp. were divided into three different morphological groups (I–III). This methodology allowed to differentiate more than 24 species of Acanthamoeba (Khan 2006; Visvesvara et al. 2007; Kłopocka et al. 2009; Fuerst et al. 2015; Derda et al. 2016). Most AK infections are caused by representatives of group II, though some isolates from group III have also been described as causative agents of the infection. Among the species of Acanthamoeba causing AK, the most prevalent are Acanthamoeba polyphaga and A. castellanii, although A. culbertsoni, A. rhysodes, A. griffini, A. quina, and A. lugdunensis have also been described as causing the infection (Clarke and Niederkorn 2006; Visvesvara 2010; Lorenzo-Morales et al. 2015).
Nevertheless, the classification based on morphologic criteria is considered ambiguous and unreliable currently. Species morphology might change depending on culture media conditions, resulting in variations in cyst morphology, an important feature for species identification. Furthermore, several studies have shown disagreements in the morphology of cysts from the same isolates, which implicates that morphological identification, alone, should not be used for species identification, requiring the use of molecular approaches (Khan 2006; Castrillón and Orozco 2013). Currently, molecular classification methods have been generated, generally classifying isolates based on the nuclear small subunit 18S ribosomal RNA whole gene sequence (Rns). This approach allows the differentiation of Acanthamoeba spp. into 22 genotypes (T1–T22) and encompasses all the Acanthamoeba isolates found so far (Corsaro et al. 2015; Fuerst et al. 2015; Corsaro et al. 2017; Taher et al. 2018).
Molecular techniques, especially the ones using ribosomal RNA (rRNA) amplification of the Rns amplicon ASA.S1 of 18S rRNA, are increasingly being used for rapid and helpful detection of Acanthamoeba. This fragment encodes the DF3 region, and the obtained sequences are compared with the Acanthamoeba reference strain sequence database. Because a reliable result is necessary, a similarity higher than 95% between isolates and reference sequences is recommended due to the high similarity between genotypes (Visvesvara et al. 2007; Lorenzo-Morales et al. 2015).
Several studies worldwide suggest that the predominant genotype in both keratitis and non-keratitis samples is the T4 genotype. Meanwhile, in a lower frequency, T2, T3, T5, T6, T8, T9, T11, T13, and T15 genotypes have also been isolated from patients with AK. Indeed, most of the genotypes known to date have been reported in human infection at least once. However, to the best of our knowledge, there is no study comprising global cases altogether. Instead, studies were limited to regional focus (Khan et al. 2002; Maghsood et al. 2005; Iovieno et al. 2010; Booton et al. 2009; Risler et al. 2013; Walochnik et al. 2014; Niyyati and Rezaeian 2015).
This systematic review proposes to assess the distribution of Acanthamoeba sp. genotypes in keratitis cases around the world. Here, we compare the frequency of AK among different geographic regions, considering the kind of sample collection and the identification method used.
Materials and methods
This systematic review was conducted by searching for articles in the English language on PubMed, Science Direct, Scielo, and Google Scholar databases. No restrictions were placed on studies’ dates, and the search returned journal articles from 2002 to 2020. The keywords used and combined in our search strategy were “Acanthamoeba,” “keratitis,” and “genotype.” All studies that assessed Acanthamoeba sp. genotypes from keratitis patients’ samples all around the world were included in this review. Studies in which samples were other than from keratitis patients, or were from contact lens apparatus, were excluded. Articles that did not specify the country where the samples were collected, or the genotype were also excluded.
All required data, such as the number of cases, type of sample collected, genotype, molecular biology method employed, primers used, pairwise sequence identities, species of Acanthamoeba, country of collection, and the year of the study, were extracted from each of the eligible articles and entered into Microsoft Excel software. It is important to note that all data presented in our research strictly reflect those reported by the original articles.
Results
A total of 2934 articles were addressed based on four databases: PubMed, Science Direct, Scielo, and Google Scholar. From those articles, 65 articles were used in the current study, as they met the previously selected inclusion criteria.
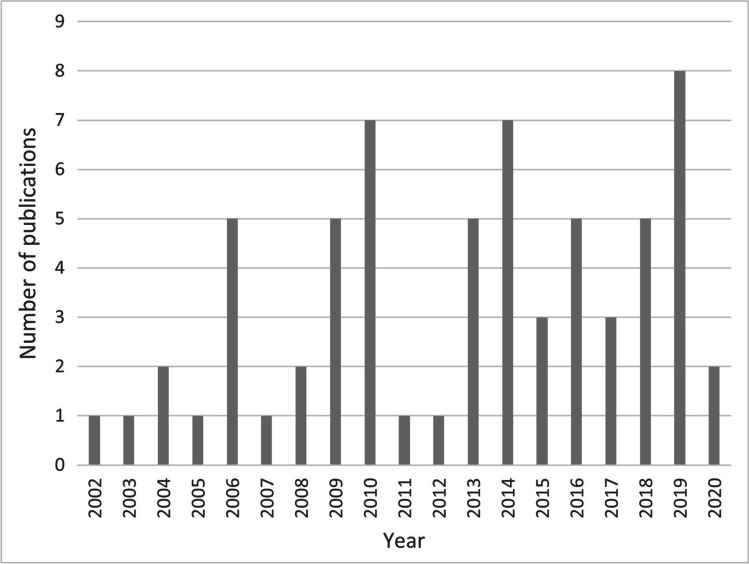
Our study collected data on the number of published articles containing cases of human keratitis caused by Acanthamoeba spp., the respective genotypes, and the year of publication. The number of publications per year can be seen in Fig. 1, where it is also pointed the first publication occurred in 2002 (Booton et al. 2002).
Fig. 1.
Publications per year of Acanthamoeba keratitis, indicating the genotype of the isolate and year of publication
Regarding the sample collected in each study, we can note a wide diverse type. Corneal scrapes were undoubtedly the most collected samples. However, most authors used more than one method for sampling in their studies. Usually, authors associate corneal scrapings with corneal biopsies, contact lenses, corneal swabs, and contact lens apparatus (lens maintenance solution, lens case). These associations can be seen in detail in Table 1.
Table 1.
Details of the collected samples in each study with their respective references
| Type of sample collected | Number of studies using this type of sample | References |
|---|---|---|
| Corneal scrapings | 29 | Sharma et al. 2004; Zhang et al. 2004; Spanakos et al. 2006; Ertabaklar et al. 2007; Ozkoc et al. 2008; Booton et al. 2009; Iovieno et al. 2010; Nagyová et al. 2010b; Niyyati et al. 2010; Nuprasert et al. 2010; Lorenzo-Morales el al. 2011; Arnalich-Montiel et al. 2013b; Duarte et al. 2013; Buerano et al. 2014; Ghamilouie et al. 2014a, b a; Ghamilouie et al. 2014a, b b; Behera et al. 2016; Padzik et al. 2016; Tawfeek et al. 2016; Alves et al. 2018; Buchele et al. 2018; Fabres et al. 2018; Taher et al. 2018; Bahreini et al. 2019; Omaña-Molina et al. 2019; Orosz et al. 2019; Tananuvat et al. 2019; Alver et al. 2020; Prithiviraj et al. 2020 |
| Corneal scrapings, contact lenses | 6 | Yera et al. 2008; Dendana et al. 2013; González-Robles et al. 2014; Hajialilo et al. 2016; Omaña-Molina et al. 2016; Casero et al. 2017 |
| Corneal scrapings, contact lenses, and contact lens paraphernalia | 6 | Booton et al. 2002; Mubareka et al. 2006; Lorenzo-Morales et al. 2007; Niyyati et al. 2009; Antonelli et al. 2018; Esboei et al. 2020 |
| Corneal scrape, biopsies and/or cotton swabs, contact lenses, and contact lens paraphernalia | 6 | De Jonckheere 2003; Di Cave et al. 2009; Ledee et al. 2009; Risler et al. 2013; Chegeni et al. 2019; Jercic et al. 2019 |
| Corneal scrapings, corneal biopsies | 4 | Maghsood et al. 2005; Yera et al. 2007; Gatti et al. 2010; Arnalich-Montiel et al, 2014 |
| Corneal scrapings and swabs, contact lens, and contact lens paraphernalia (lens case, lens solutions) | 3 | Abe and Kimata 2010; Di Cave et al. 2014; Wagner et al. 2016 |
| Symptomatic keratitis human patients, corneal samples | 3 | Rahman et al. 2013; Rocha-Cabrera et al. 2015; Martín-Pérez et al. 2017 |
| Contact lens | 1 | Heredero-Bermejo et al. 2015 |
| Corneal scraping, contact lens, and amniotic membrane | 1 | Sharifi et al. 2010 |
| Corneal scrapings and corneal button | 1 | Zhao et al. 2010 |
In addition, only one study used unconventional samples, such as corneal button (Zhao et al. 2010) and amniotic membrane, a graft used to treat corneal epithelial defects (Sharifi et al. 2010). Some studies did not specify what kind of sample was collected, using only terms such as “symptomatic keratitis human patients” or “corneal samples.”
In terms of molecular approaches, we can notice that many studies used the conventional PCR technique to detect and identify Acanthamoeba isolates. This technique was used in combination with other molecular methods for quantification or phylogenetic analysis (Ghamilouie et al. 2014b; Antonelli et al. 2018; Orosz et al. 2018). The most widely used set of primers in the studies was JDP1 and JDP2, being present in 15 studies. These primers allow the amplification of the Rns amplicon ASA.S1 from 18S rRNA, which encodes the highly variable DF3 region.
For a reliable genotyping of Acanthamoeba isolates, amplification and sequencing of the target gene are necessary. Regarding the similarity amidst the genotypes belonging to the database, most of the articles showed data with similarity > 95%. However, there were found articles that did not bring this information in their results.
In the returned studies, genotyped AK cases were found in 4 continents: Asia, America, Europe, and Africa. Seven in Asia and America, 11 in Europe, and two in Africa, a total of 27 countries around the world. In addition, two of the studies did not specify the country where the samples were collected. Regardless, these studies were included in the systematic review since it was possible to know the continent to which they belonged, “North America,” and “Southern Africa.” No studies from the Oceania continent have been found so far.
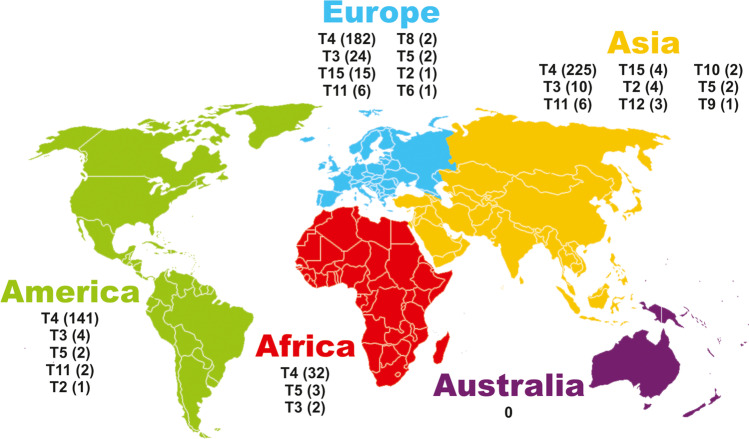
A total of 675 cases of amoebic keratitis caused by Acanthamoeba spp. were found to be genotyped worldwide. From those cases, 253 were from Asia, 150 from America, 233 from Europe, and 40 from Africa. The total number of cases per genotype in each continent can be seen in Fig. 2. In this same figure, we can see that the T4 genotype was the most prevalent, and Asia was the continent with more cases of this genotype.
Fig. 2.
Relation between AK cases with the continents and genotypes returned. The number of cases of each genotype can be seen in parentheses
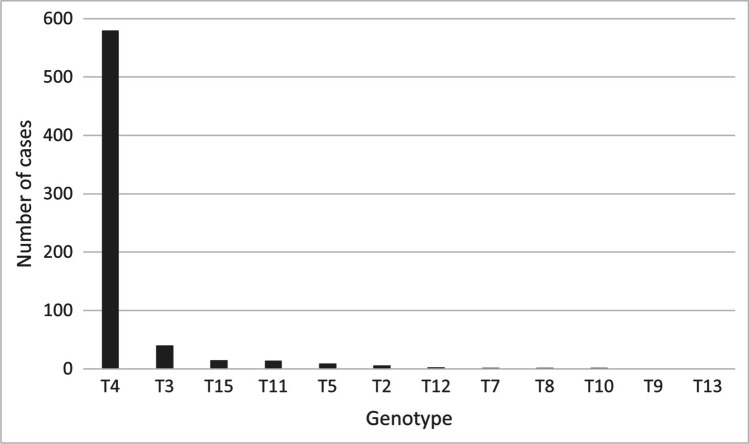
In Fig. 3, it is evident that the T4 genotype is the most prevalent worldwide, corresponding to 580 of the total 675 cases. The frequencies of the other genotypes were T3 (40), T15 (15), T11 (14), T5 (9), T2 (6), T12 (3), T7 (2), T8 (2), T10 (2), T9 (1), and T13 (1). When it comes to percentages, the prevalence of each genotype was: T4 (85.92%), T3 (5.92%), T15 (2.22%), T11 (2.07%), T5 (1.33%), T2 (0.88%), T12 (0.44%), T7 (0.29%), T8 (0.29%), T10 (0.29%), T9 (0.15%), and T13 (0.15%). In addition, it is important to note that in the cases described in this systematic review, the genotypes T1, T6, T14, T16, T17, T18, T19, T20, T21, and T22 were not found causing AK.
Fig. 3.
Worldwide cases of Acanthamoeba keratitis and their respective genotypes
Table 2 shows, with more details, the number of cases for each genotype among countries, along with the respective references.
Table 2.
Returned genotypes by country where the sample comes from. The number of cases for each genotype can be found in parentheses
| Continent | Genotypes | References |
|---|---|---|
| Asia | ||
| China | T4 (55) T3 (2) | Booton et al. 2002; Zhang et al. 2004; Zhao et al. 2010; Li et al. 2019 |
| India | T4 (56), T11 (1), T12 (3), T10 (2) | Sharma et al. 2004; Behera et al. 2016; Prithiviraj et al. 2020 |
| Iran |
T4 (80), T3 (5), T11 (5), T2 (3), T9 (1) |
Niyyati et al. 2009; Niyyati et al. 2010; Ghamilouie et al. 2014a; Ghamilouie et al. 2014b; Maghsood et al. 2015; Hajialilo et al. 2016; Bahreini et al. 2019; Chegeni et al. 2019; Esboei et al. 2020 |
| Japan | T4 (26), T3 (3), T5 (1) | Abe and Kimata 2010; Rahman et al. 2013 |
| Philippines | T4 (1) | Buerano et al. 2014 |
| Thailand | T4 (5), T5 (1) | Nuprasert et al. 2010; Tananuvat et al. 2019 |
| Turkey | T4 (2), T2 (1) | Ertabaklar et al. 2007; Ozkoc et al. 2008; Alver et al. 2020 |
| America | ||
| Argentina | T4 (10) | Casero et al. 2017 |
| Brazil | T4 (7) | Duarte et al. 2013; Alves et al. 2018; Buchele et al.2018; Fabres et al. 2018 |
| Canada | T4 (1) | Mubareka et al. 2006 |
| Chile | T4 (73), T11 (2), T2 (1) | Jercic et al. 2019 |
| Mexico | T4 (2), T3 (2) | González-Robles et al. 2014; Omaña-Molina et al. 2016; Omaña-Molina et al. 2019; |
| USA | T4 (35), T3 (2), T5 (1) | Booton et al. 2009, Ledee et al. 2009 |
| Venezuela | T4 (13) | Wagner et al. 2016 |
| North America | T5 (1) | Iovieno et al. 2010 |
| Europe | ||
| Austria | T6 (1) | Blaschitz et al. 2006 |
| Belgium | T4 (15) | De Jonckheere 2003 |
| Czech Republic | T3 (1) | Nagyová et al. 2010b |
| France | T4 (33), T3 (3), T2 (1), T5 (1), T11 (1) | Yera et al. 2007; Yera et al. 2008; Risler et al. 2013 |
| Greece | T4 (4), T5 (1) | Spanakos et al. 2006 |
| Hungary | T4 (6), T8 (2) | Orosz et al. 2018; Orosz et al. 2019 |
| Italy | T4 (65), T15 (13), T3 (12), T11 (1) | Di Cave et al. 2009; Del Gatti et al. 2010; Di Cave et al. 2014; Antonelli et al. 2018 |
| Poland | T4 (6) | Padzik et al. 2016 |
| Slovakia | T4 (3), T15 (1) | Nagyová et al. 2010b |
| Spain | T4 (40), T3 (7), T11 (3) | Lorenzo-Morales et al. 2007; Lorenzo-Morales el al. 2011; Arnalich-Montiel et al. 2013a; Arnalich-Montiel et al. 2013b; Arnalich-Montiel et al, 2014; Heredero-Bermejo et al. 2015; Rocha-Cabrera et al. 2015; Martín-Pérez et al. 2017 |
| Sweden | T4 (10), T3 (1), T11 (1), T15 (1) | Sharifi et al. 2010 |
| Africa | ||
| Egypt | T4 (27), T5 (3), T3 (2), T7 (2) | Tawfeek et al. 2016; Taher et al. 2018 |
| Tunisia | T4 (5) | Dendana et al. 2013 |
| Southern Africa | T13 (1) | Grün et al. 2014 |
This systematic review showed that the T4 genotype, in addition to being responsible for the largest number of cases worldwide, also contains the larger number of Acanthamoeba species. In addition, it can be seen in Table 3 that the same species is related to different genotypes; for example, both A. castellani and A. palestinensis belong to genotype T4 and T2, A. culbertsoni belongs to genotype T4 and T10, and A. hatchetti belongs to genotype T4, T6, and T11. However, it is important to note that most of the articles do not have the identification of Acanthamoeba species.
Table 3.
Acanthamoeba species related to each genotype and their respective references
| Genotypes | Species | References |
|---|---|---|
| T4 | A. castellani, A. polyphaga, A. palestinensis, A. culbertsoni, A. triangularis, A. rhysodes, A. royreba, A. quina, A. hatchetti | Maghsood et al. 2005; Spanakos et al. 2006; Ertabaklar et al. 2007; Ozkoc et al. 2008; Di Cave et al. 2009; Sharifi et al. 2010; Di Cave et al. 2014; Ghamilouie et al. 2014b; Omaña-Molina et al. 2016; Casero et al. 2017; Taher et al. 2018; Omaña-Molina et al. 2019; Prithiviraj et al. 2020 |
| T2 | A. castellani; A. palestinensis | Maghsood et al. 2005; Alver et al. 2020 |
| T3 | A. griffini | Maghsood et al. 2005; Sharifi et al. 2010; Di Cave et al. 2014; González-Robles et al. 2014; Heredero-Bermejo et al. 2015; Omaña-Molina et al. 2016; Taher et al. 2018 |
| T5 | A. lenticulata | Spanakos et al. 2006; Ledee et al. 2009; Iovieno et al. 2010; Rahman et al. 2013; Taher et al. 2018 |
| T6 | A. hatchetti | Blaschitz et al. 2006 |
| T7 | A. astronyxis | Tawfeek et al. 2016 |
| T11 | A. stevensoni, A. hatchetti | Sharifi et al. 2010; Lorenzo-Morales el al. 2011; Hajialilo et al. 2016; Prithiviraj et al. 2020 |
| T10 | A. culbertsoni | Behera et al. 2016 |
| T15 | A. jacobsi | Di Cave et al. 2009; Sharifi et al. 2010; Di Cave et al. 2014 |
Discussion
As a result of its ubiquity, the cosmopolitan Acanthamoeba protozoan poses a risk to human health due to its ability to infect the host and survive in the environment. Studies are still needed to clarify the pathogenicity of AK and other infections caused by this free-living amoeba, which can help enhance the search for therapeutic targets (Ledee et al. 2009).
Our study shows that in the last 20 years there has been a gradual increase in the number of studies genotyping Acanthamoeba isolates causing keratitis, especially in the last decade. Unfortunately, the pandemic of Sars-Cov-2 caused the number of publications to decrease to a similar number as 20 years ago, which is certainly an exception to the upward trend. Several factors are related to the increase in the number of AK cases worldwide every year, such as the widespread use of contact lenses for vision correction or cosmetic purposes together with better diagnostics, therefore becoming classified as an emerging disease (Panjwan 2010; Astorga et al. 2011). However, we have reasons to believe that this number is still underreported and, in addition, not all diagnosed cases are genotyped. Although there are several genotyping studies in some countries, to the best of our knowledge, there is no study listing all AK cases, thus providing a comprehensive view of the frequency of AK genotypes worldwide.
Different sample collection methods have been shown to be effective for isolating Acanthamoeba spp. in keratitis. Among these methods, there are corneal scraping, corneal biopsies, and corneal smears, as well as collections of contact lenses and their accessories, like lens cases and cleaning solutions. However, we can note that a predominant part of the studies chose to use corneal scrapes and contact lenses from AK patients.
It is recommended that the detection of Acanthamoeba spp. in keratitis patients be performed by molecular tests, such as conventional PCR or real-time PCR, with the last being faster and more accurate than the first one (Visvesvara et al. 2007; Corsaro et al. 2015; Maycock and Jayaswal 2016). Furthermore, in one study, Sharma et al. (2004) used a real-time multiplex PCR assay to detect Acanthamoeba, a molecular method that proved to be reliable and with sufficient sensitivity to detect up to 5 picograms of Acanthamoeba DNA.
When analyzing all genotyped cases of AK, our study shows that there were 580 cases genotyped as T4, corresponding to 85.92% of the total number of cases reported worldwide. In addition, the T4 genotype was the most prevalent on all continents where AK was found, suggesting that even in different countries, isolates with this genotype may have similar pathogenic properties. It is known that an important initial step in the pathology of AK is the adherence to corneal epithelial cells, which is strongly related to the expression of mannose-binding protein by Acanthamoeba sp. In the T4 genotype, this mannose-binding protein appears to bind more firmly to the membrane surface of host cells, which makes this genotype more cytotoxic than the others, and eventually being responsible for a higher number of infections (Hurt et al. 2003; Garate et al. 2006; Ledee et al. 2009; Noorjahan 2010). Furthermore, the exposure to mannose leads to the liberation of a low molecular weight protease called MIP133, which can have a cytolytic effect on corneal epithelial cells (Hurt et al. 2003; Garate et al. 2006; Ledee et al. 2009; Noorjahan 2010). As a result, it is assumed that T4 is the most virulent genotype, having properties that enhance its transmissibility, given its greatest environmental distribution (Maghsood et al. 2005; Ledee et al. 2009). Therefore, the highest expression of mannose-binding protein in the T4 genotype could be an effective target for new therapeutic approaches that would serve as a treatment for the vast majority of cases of amoebic keratitis.
We can also note that the genotype distribution among the four continents is very similar. In addition to T4, our study shows that the second most prevalent genotype was T3. It was the second most prevalent genotype in three of the four continents. The exception occurred in Africa, where the T5 genotype was the second most prevalent. However, this difference is not significant since the T5 genotype has only one more case than T3 in that continent. The T3 genotype was present in 40 reported cases, followed by T15 with 15 cases, T11 with 14, and T5 with 9. Other genotypes were also reported causing keratitis, although less frequently, as T2 (6 cases), T12 (3 cases), T7, T8, and T10 (2 cases each). Furthermore, T9 and T13 genotypes also caused Acanthamoeba keratitis, with one case each. However, it is important to note that the diagnosis of AK is complex, and the larger part of diagnosed cases were not genotyped. Furthermore, it is evident in this study that there is a difference in the number of genotyped cases among the continents, with the African continent having the lowest number of cases and Asia the largest, likely on account of support resources available. Therefore, our study describes the genotypic distribution of cases reported so far worldwide.
According to the reported scientific literature, infections caused by non-T4 genotypes are more aggressive, and the outcomes are extremely unfavorable, although the T4 genotype is the most prevalent (Iovieno et al. 2010; Sharifi et al. 2010). Because of this, we believe that the ideal scenario would consist of genotyping all isolates, although we know that it is almost impossible in practice.
In addition, reports of resistance to multipurpose cleaning solutions have already been related to non-T4 genotypes, like T3 and T5 (Shoff et al. 2007). Although multipurpose cleaning solutions are composed of different molecules from the medicines used for AK treatments, it indicates that such genotypes are more resistant, which can be related to a poor response to medical therapy. Moreover, according to Arnalich-Montiel et al. (2014), non-T4 genotypes have longer delays to diagnosis when compared with T4 genotypes and greater need for surgical intervention, that is, worse clinical outcomes.
Our study makes clear that, though A. castellani and A. polyphaga are the species most frequent in the T4 genotype, other species can also belong to this genotype, such as A. culbertsoni, A. triangularis, A. rhysodes, A. royreba, A. quina, and A. hatchetti. In addition, A. castellani and A. palestinensis were also related to the T2 genotype, and the T3 genotype was only related to A. griffini. It is clear that the classification of Acanthamoeba into species and genotypes still needs to be improved to clearly understand this relation. Furthermore, we agree with a recent study from Corsaro et al. (2020), which states that the identification of Acanthamoeba should currently have a more transparent organization than the classification based only on morphology that once was appropriate. Still today, it is a cause of confusion, relating several species to the same genotype.
It is important to highlight that there is not yet any effective treatment that could be used in all cases of AK. Therefore, novel therapeutics are needed to eliminate both amoeba life forms: trophozoites and cysts. It is important to remember that cyst forms are very resistant and related to the recurrence of infection. Given the differences in the pathology, virulence, susceptibility to treatment, and clinical outcomes between genotypes, genotype isolates are a path to be taken to have a better correlation between in vitro and in vivo efficacies, resulting in more effective therapies and successful treatment in AK cases.
In summary, Acanthamoeba genotyping has great importance for taxonomic purposes, understanding the geographical distribution of species, and also for clinical and epidemiological studies to better clarify this infection’s pathology and clinical outcomes. The literature shows that non-T4 genotypes can lead to more severe symptoms and have poorer response to medical therapy than genotype T4 (Arnalich-Montiel et al. 2014), though more than 85% of Acanthamoeba keratitis cases have been linked with T4 genotype, which is why it is supposedly the most virulent. AK remains a challenging disease to diagnose and treat, and further studies should be conducted to elucidate what makes some genotypes more pathogenic than others. This information would also play a fundamental role in providing new reliable diagnosis methods and novel therapeutic strategies.
Acknowledgements
The authors thank the Department of Microbiology, Immunology and Parasitology of Federal University of Rio Grande do Sul and CNPq.
Funding
This study received financial support from the National Council for Scientific and Technological Development (CNPq).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abe N, Kimata I. Genotyping of Acanthamoeba isolates from corneal scrapings and contact lens cases of Acanthamoeba keratitis patients in Osaka. Japan Jpn J Infect Dis. 2010;63(4):299–301. [PubMed] [Google Scholar]
- Alver O, Baykara M, YÜrÜk M, TÜzemen NÜ. Acanthamoeba keratitis and Acanthamoeba conjunctivitis: a case report. Iran J Parasitol. 2020;15(2):272–277. [PMC free article] [PubMed] [Google Scholar]
- Alves DSMM, Gonçalves GS, Moraes AS, Alves LM, Neto JRC, Hecht MM, Nitz N, Gurgel-Gonçalves R, Bernardes G, Castro AM, Chalita MR, Vinaud MC. The first Acanthamoeba keratitis case in the Midwest region of Brazil: diagnosis, genotyping of the parasite and disease outcome. Rev Soc Bras Med Trop. 2018;51(5):716–719. doi: 10.1590/0037-8682-0010-2018. [DOI] [PubMed] [Google Scholar]
- Antonelli A, Favuzza E, Galano A, Di Filippo MM, Ciccone N, Berrilli F, Mencucci R, Di Cave D, Rossolini GM. Regional spread of contact lens-related Acanthamoeba keratitis infection in Italy. New Microbiol. 2018;41(1):83–85. [PubMed] [Google Scholar]
- Arnalich-Montiel F, Jaumandreu L, Leal M, Valladares B, Lorenzo-Morales J. Scleral and intraocular amoebic dissemination in Acanthamoeba keratitis. Cornea. 2013;32(12):1625–1627. doi: 10.1097/ICO.0b013e31829ded51. [DOI] [PubMed] [Google Scholar]
- Arnalich-Montiel F, Lorenzo-Morales J, Irigoyen C, Morcillo-Laiz R, López-Vélez R, Muñoz-Negrete F, Piñero JE, Valladares B. Co-isolation of Vahlkampfia and Acanthamoeba in Acanthamoeba-like keratitis in a Spanish population. Cornea. 2013;32(5):608–614. doi: 10.1097/ICO.0b013e31825697e6. [DOI] [PubMed] [Google Scholar]
- Arnalich-Montiel F, Lumbreras-Fernández B, Martín-Navarro CM, Valladares B, Lopez-Velez R, Morcillo-Laiz R, Lorenzo-Morales J. Influence of Acanthamoeba genotype on clinical course and outcomes for patients with Acanthamoeba keratitis in Spain. J Clin Microbiol. 2014;52:1213–1216. doi: 10.1128/jcm.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astorga B, Lorenzo-Morales J, Martín-Navarro CM, Alarcón V, Moreno J, González AC, Navarrete E, Piñero JE, Valladares B. Acanthamoeba belonging to T3, T4, and T11: genotypes isolated from air-conditioning units in Santiago, Chile. J Eukaryot Microbiol. 2011;58:542–544. doi: 10.1111/j.1550-7408.2011.00584.x. [DOI] [PubMed] [Google Scholar]
- Bahreini MS, Motazedian MH, Bamdad S, Afshar MJA, Asgari QQ (2019) Detection of free living amoeba infection in patients with suspected central nervous system and keratitis disease in Shiraz, Southern Iran. J Health Sci Surveillance 7:2. 10.30476/JHSSS.2020.84999.1052
- Behera HS, Panda A, Satpathy G, Bandivadekar P, Vanathi M, Agarwal T, Nayak N, Tandon R (2016) Genotyping of Acanthamoeba spp. and characterization of the prevalent T4 type along with T10 and unassigned genotypes from amoebic keratitis patients in India. J Med Microbiol 65(5):370–376. 10.1099/jmm.0.000234 [DOI] [PubMed]
- Berger P, Papazian L, Drancourt M, La Scola B, Auffray JP, Raoult D. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg Infect Dis. 2006;12:248–255. doi: 10.3201/eid1202.050434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschitz M, Köhsler M, Aspöck H, Walochnik J. Detection of a serine proteinase gene in Acanthamoeba genotype T6 (Amoebozoa: Lobosea) Exp Parasitol. 2006;114(1):26–33. doi: 10.1016/j.exppara.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Booton GC, Joslin CE, Shoff M, Tu EY, Kelly DJ, Fuerst PA (2009) Genotypic identification of Acanthamoeba sp. isolates associated with an outbreak of Acanthamoeba keratitis. Cornea 28(6):673–676. 10.1097/ICO.0b013e31819342a7 [DOI] [PMC free article] [PubMed]
- Booton GC, Kelly DJ, Chu YW, Seal DV, Houang E, Lam DS, Byers TJ, Fuerst PA. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J Clin Microbiol. 2002;40:1621–1625. doi: 10.1128/jcm.40.5.1621-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booton GC, Rogerson A, Bonilla TD, Seal DV, Kelly DJ, Beattie TK, Tomlinson A, Lares-Villa F, Fuerst PA, Byers TJ (2004) Molecular and physiological evaluation of subtropical environmental isolates of Acanthamoeba spp., causal agent of Acanthamoeba keratitis. J Eukaryot Microbiol 51(2):192–200. 10.1111/j.1550-7408.2004.tb00545.x [DOI] [PubMed]
- Buchele MLC, Wopereis DB, Casara F, Macedo JP, Rott MB, Monteiro FBF, Bazzo ML, Spada FR, Santos JI, Caumo KS. Contact lens-related polymicrobial keratitis: Acanthamoeba spp. genotype T4 and Candida albicans. Parasitol Res. 2018;117:3431–3436. doi: 10.1007/s00436-018-6037-x. [DOI] [PubMed] [Google Scholar]
- Buerano CC, Trinidad AD, Fajardo LS, Cua IY, Baclig MO, Natividad FF. Isolation of acanthamoeba genotype t4 from a non-contact lens wearer from the Philippines. Trop Med Health. 2014;42(4):145–147. doi: 10.2149/tmh.2014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RD, Mongi F, Laconte L, Rivero F, Sastre D, Teherán A, Herrera G, Ramírez JD. Molecular and morphological characterization of Acanthamoeba isolated from corneal scrapes and contact lens wearers in Argentina. Infect Genet Evol. 2017;54:170–175. doi: 10.1016/j.meegid.2017.06.031. [DOI] [PubMed] [Google Scholar]
- Castrillón JC, Orozco LP. Acanthamoeba spp. como parásitos patógenos y oportunistas. Rev Chilena Infectol. 2013;30(2):147–155. doi: 10.4067/S0716-10182013000200005. [DOI] [PubMed] [Google Scholar]
- Chegeni TN, Ghaffarifar F, Pirestani M, Khoshzaban F, Dalimi A, Maspi N. Genotyping of Acanthamoeba species isolated from keratitis patients by PCR sequencing methods in Tehran, Iran. Int J Med Lab. 2019;6(4):259–265. [Google Scholar]
- Clarke DW, Niederkorn JY. The pathophysiology of Acanthamoeba keratitis. Trends Parasitol. 2006;22:175–180. doi: 10.1016/j.pt.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Köhsler M, Filippo MM, Venditti D, Monno R, Di Cave D, Berrilli F, Walochnik J. Update on Acanthamoeba jacobsi genotype T15, including full-length 18S rDNA molecular phylogeny. Parasitol Res. 2017;116:1273–1284. doi: 10.1007/s00436-017-5406-1. [DOI] [PubMed] [Google Scholar]
- Corsaro D. Update on Acanthamoeba phylogeny. Parasitol Res. 2020;119:3327–3338. doi: 10.1007/s00436-020-06843-9. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Walochnik J, Köhsler M, Rott MB. Acanthamoeba misidentification and multiple labels: redefining genotypes T16, T19, and T20 and proposal for Acanthamoeba micheli sp. nov. (genotype T19) Parasitol Res. 2015;114:2481–2490. doi: 10.1007/s00436-015-4445-8. [DOI] [PubMed] [Google Scholar]
- Costa AO, Castro EA, Ferreira GA, Furst C, Crozeta MA, Thomaz-Soccol V. Characterization of Acanthamoeba isolates from dust of a public hospital in Curitiba, Paraná, Brazil. J Eukaryot Microbiol. 2010;57:70–75. doi: 10.1111/j.1550-7408.2009.00453.x. [DOI] [PubMed] [Google Scholar]
- Dart JK, Saw VP, Kilvington S. Acanthamoeba keratitis: diagnosis and treatment update 2009. Am J Ophthalmol. 2009;148:487–499. doi: 10.1016/j.ajo.2009.06.009. [DOI] [PubMed] [Google Scholar]
- De Jonckheere JF. Epidemiological typing of Acanthamoeba strains isolated from keratitis cases in Belgium. Bull Soc Belge Ophtalmol. 2003;287:27–33. [PubMed] [Google Scholar]
- Dendana F, Sellami H, Trabelsi H, Neji S, Cheikhrouhou F, Makni F, Ayadi A. Acanthamoeba T4 genotype associated with keratitis infections in Tunisia. Parasitol Res. 2013;112(1):401–405. doi: 10.1007/s00436-012-3149-6. [DOI] [PubMed] [Google Scholar]
- Derda M, Wojtkowiak-Giera A, Kolasa-Wołosiuk A, Kosik-Bogacka D, Hadaś E, Jagodziński PP, Wandurska-Nowak E. Acanthamoeba infection in lungs of mice expressed by toll-like receptors (TLR2 and TLR4) Exp Parasitol. 2016;165:30–34. doi: 10.1016/j.exppara.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Diaz JH. Increasing intracerebral infections caused by free-living amebae in the United States and Worldwide. J of Neuroparasitol. 2010;1:104. doi: 10.4303/jnp/N100801. [DOI] [Google Scholar]
- Di Cave D, D’ Alfonso R, Comlavi KAD, D’ Orazi C, Monno R, Berrilli F (2014) Genotypic heterogeneity based on 18S-rRNA gene sequences among Acanthamoeba isolates from clinical samples in Italy. Exp Parasitol 145Suppl:S46-S49. 0.1016/j.exppara.2014.05.009 [DOI] [PubMed]
- Di Cave D, Monno R, Bottalico P, Guerriero S, D’Amelio S, D’Orazi C, Berrilli F. Acanthamoeba T4 and T15 genotypes associated with keratitis infections in Italy. Eur J Clin Microbiol Infect Dis. 2009;28(6):607–612. doi: 10.1007/s10096-008-0682-4. [DOI] [PubMed] [Google Scholar]
- Duarte JL, Furst C, Klisiowicz DR, Klassen G, Costa AO. Morphological, genotypic, and physiological characterization of Acanthamoeba isolates from keratitis patients and the domestic environment in Vitoria, Espírito Santo. Brazil Exp Parasitol. 2013;135(1):9–14. doi: 10.1016/j.exppara.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Ertabaklar H, Türk M, Dayanir V, Ertuğ S, Walochnik J. Acanthamoeba keratitis due to Acanthamoeba genotype T4 in a non-contact-lens wearer in Turkey. Parasitol Res. 2007;100(2):241–246. doi: 10.1007/s00436-006-0274-0. [DOI] [PubMed] [Google Scholar]
- Esboei BR, Fakhar M, Saberi R, Baratif M, Moslemig M, Hassannia H, Moghadame Y, Jalalloua N. Genotyping and phylogenic study of Acanthamoeba isolates from human keratitis and swimming pool water samples in Iran. Parasite Epidemiol Control. 2020;11:00164. doi: 10.1016/j.parepi.2020.e00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabres LF, Maschio VJ, Santos DLD, Kwitko S, Ruschel Marinho DR, Araújo BS, Locatelli CI, Rott MB. Virulent T4 Acanthamoeba causing keratitis in a patient after swimming while wearing contact lenses in Southern Brazil. Acta Parasitol. 2018;63(2):428–432. doi: 10.1515/ap-2018-0050. [DOI] [PubMed] [Google Scholar]
- Fraser MN, Wong Q, Shah L, Holland SP, Morshed M, Isaac-Renton J, Chong M, Kibsey P, Patrick DM. Characteristics of an Acanthamoeba keratitis outbreak in British Columbia between 2003 and 2007. Ophthalmology. 2012;119:1120–1125. doi: 10.1016/j.ophtha.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Fuerst PA, Booton GC, Crary M. Phylogenetic analysis and the evolution of the 18S rRNA gene typing system of Acanthamoeba. J Eukaryot Microbiol. 2015;62:69–84. doi: 10.1111/jeu.12186. [DOI] [PubMed] [Google Scholar]
- Garate M, Marchant J, Cubillos I, Cao Z, Khan NA, Panjwani N. In vitro pathogenicity of Acanthamoeba is associated with the expression of the mannose-binding protein. Invest Ophthalmol vis Sci. 2006;47:1056–1062. doi: 10.1167/iovs.05-0477. [DOI] [PubMed] [Google Scholar]
- Gatti S, Rama P, Matuska S, Berrilli F, Cavallero A, Carletti S, Bruno A, Maserati R, Di Cave D. Isolation and genotyping of Acanthamoeba strains from corneal infections in Italy. J Med Microbiol. 2010;59:1324–1330. doi: 10.1099/jmm.0.019786-0. [DOI] [PubMed] [Google Scholar]
- Ghamilouie MM, Valadkhani Z, Khoshzaban F. Species identification of Acanthamoeba strains isolated from patients referring to Farabi Eye Reference Center using PCR-RFLP method. J Med Microbiol Infec Dis. 2014;2(3):125–129. [Google Scholar]
- Ghamilouie MM, Valadkhani Z, Rahimi F, Khoshzaban F, Aghighi Z, Hassan N. Isolation and genotyping of Acanthamoeba strains from corneal scraps. Iranian J Ophthalmol. 2014;26(2):97–101. [Google Scholar]
- Gokhale NS. Medical management approach to infectious keratitis. Indian J Ophthalmol. 2008;56:215–220. doi: 10.4103/0301-4738.40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Robles A, Salazar-Villatoro L, Omaña-Molina M, Reyes-Batlle M, Martín-Navarro CM, Lorenzo-Morales J. Morphological features and in vitro cytopathic effect of Acanthamoeba griffini trophozoites isolated from a clinical case. J Parasitol Res. 2014;2014:10. doi: 10.1155/2014/256310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–433. doi: 10.1128/cmr.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün AL, Stemplewitz B, Scheid P. First report of an Acanthamoeba genotype T13 isolate as etiological agent of a keratitis in humans. Parasitol Res. 2014;113(6):2395–2400. doi: 10.1007/s00436-014-3918-5. [DOI] [PubMed] [Google Scholar]
- Hajialilo E, Behnia M, Tarighi F, Niyyati M, Rezaeian M. Isolation and genotyping of Acanthamoeba strains (T4, T9, and T11) from amoebic keratitis patients in Iran. Parasitol Res. 2016;115(8):3147–3151. doi: 10.1007/s00436-016-5072-8. [DOI] [PubMed] [Google Scholar]
- Hammersmith KM. Diagnosis and management of Acanthamoeba keratitis. Curr Opin Ophthalmol. 2006;17:327–331. doi: 10.1097/01.icu.0000233949.56229.7d. [DOI] [PubMed] [Google Scholar]
- Heredero-Bermejo I, Criado-Fornelio A, De Fuentes I, Soliveri J, Copa-Patiño JL, Pérez-Serrano J. Characterization of a human-pathogenic Acanthamoeba griffini isolated from a contact lens-wearing keratitis patient in Spain. Parasitology. 2015;142(2):363–373. doi: 10.1017/S0031182014001140. [DOI] [PubMed] [Google Scholar]
- Hurt M, Neelam S, Niederkorn J, Alizadeh H. Pathogenic Acanthamoeba spp. secrete a mannose-induced cytolytic protein that correlates with the ability to cause disease. Infect Immun. 2003;71:6243–6255. doi: 10.1128/IAI.71.11.6243-6255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovieno A, Oechslerb RA, Ledee DR, Miller D, Alfonso EC. Drug-resistant severe Acanthamoeba keratitis caused by rare T5 Acanthamoeba genotype. Eye Contact Lens. 2010;36:183–184. doi: 10.1097/icl.0b013e3181da2350. [DOI] [PubMed] [Google Scholar]
- Jercic MI, Aguayo C, Saldarriaga-Córdoba M, Muiño L, Chenet SM, Lagos J, Osuna A, Fernández J. Genotypic diversity of Acanthamoeba strains isolated from Chilean patients with Acanthamoeba keratitis. Parasit Vectors. 2019;12:58. doi: 10.1186/s13071-019-3302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Sun X, Wang Z, Zhang Y. Acanthamoeba keratitis: clinical characteristics and management. Ophthalmology. 2006;113:412–416. doi: 10.1016/j.jtos.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Juárez MM, Tártara LI, Cida AG, Reald JP, Bermúdeza JM, Rajala VB, Palmad SD. Acanthamoeba in the eye, can the parasite hide even more? Latest developments on the disease. Cont Lens Anterior Eye. 2018;41:245–251. doi: 10.1016/j.clae.2017.12.017. [DOI] [PubMed] [Google Scholar]
- Khan NA. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev. 2006;30:564–595. doi: 10.1111/j.1574-6976.2006.00023.x. [DOI] [PubMed] [Google Scholar]
- Khan NA, Jarroll EL, Paget TA. Molecular and physiological differentiation between pathogenic and nonpathogenic Acanthamoeba. Curr Microbiol. 2002;45:197–202. doi: 10.1007/s00284-001-0108-3. [DOI] [PubMed] [Google Scholar]
- Khan NA, Tareen NK. Genotypic, phenotypic, biochemical, physiological and pathogenicity-based categorisation of Acanthamoeba strains. Folia Parasitol. 2003;50:97–104. doi: 10.14411/fp.2003.017. [DOI] [PubMed] [Google Scholar]
- Khezri A, Fallah E, Mostafazadeh M, Spotin A, Shahbazi A, Mahami-Oskouei M, Hazratian T. Molecular and morphometric characterization of Acanthamoeba spp. from different water sources of northwest Iran as a neglected focus, co-bordeed with the country of Iraq. Jundishapur J Microbiol. 2016;9:38481. doi: 10.5812/jjm.38481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilvington S, Gray T, Dart J, Morlet N, Beeching JR, Frazer DG, Matheson M. Acanthamoeba keratitis: the role of domestic tap water contamination in the United Kingdom. Invest Ophthalmol vis Sci. 2004;45:165–169. doi: 10.1167/iovs.03-0559. [DOI] [PubMed] [Google Scholar]
- Kitzmann AS, Goins KM, Sutphin JE, Wagoner MD. Keratoplasty for treatment of Acanthamoeba keratitis. Ophthalmology. 2009;116:864–869. doi: 10.1016/j.ophtha.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Kłopocka W, Rędowicz MJ, Wasik A. Regulation of cortical cytoskeleton dynamics during migration of free-living amoebae. Postępy Biochemii. 2009;55:129–137. [PubMed] [Google Scholar]
- Kong HH, Shin JY, Yu HS, Kim J, Hahn TW, Hahn YH, Chung D. Mitochondrial DNA restriction fragment length polymorphism (RFLP) and 18S small-subunit ribosomal DNA PCR-RFLP analyses of Acanthamoeba isolated from contact lens storage cases of residents in southwestern Korea. J Clin Microbiol. 2002;40:1199–1206. doi: 10.1128/jcm.40.4.1199-1206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshler M, Leitner B, Blaschitz M, Michel R, Aspock H, Walochnik J. ITS1 sequence variabilities correlate with 18S rDNA sequence types in the genus Acanthamoeba (Protozoa: Amoebozoa) Parasitol Res. 2006;98:86–93. doi: 10.1007/s00436-005-0022-x. [DOI] [PubMed] [Google Scholar]
- Kot K, Łanocha-Arendarczyk NA, Kosik-Bogacka DI (2018) Amoebas from the genus Acanthamoeba and their pathogenic properties. Ann of Parasitol 64(4):299–308. 10.17420/ap6404.164 [PubMed]
- Król-Turmińska K, Olender A. Human infections caused by free-living amoebae. Ann Agric Environ Med. 2017;24:254–260. doi: 10.5604/12321966.1233568. [DOI] [PubMed] [Google Scholar]
- Kumar R, Lloyd D. Recent advances in the treatment of Acanthamoeba keratitis. Clin Infect Dis. 2002;35:434–441. doi: 10.1086/341487. [DOI] [PubMed] [Google Scholar]
- Lanocha N, Kosik-Bogacka D, Maciejewska A, Sawczuk M, Wilk A, Kuźna-Grygiel W. The occurence Acanthamoeba (free living amoeba) in environmental and respiratory samples in Poland. Act Protozool. 2009;48:271–279. [Google Scholar]
- Lass A, Guerrero M, Li X, Karanis G, Ma L, Karanis P. Detection of Acanthamoeba spp. in water samples collected from natural water reservoirs, sewages, and pharmaceutical factory drains using LAMP and PCR in China. Sci Total Environ. 2017;584:489–494. doi: 10.1016/j.scitotenv.2017.01.046. [DOI] [PubMed] [Google Scholar]
- Ledee DR, Booton GC, Awwad MH, Sharma S, Aggarwal RK, Nisz IA, Markus MB, Fuerst PA, Byers TJ. Advantages of using mitochondrial 16S rDNA sequences to classify clinical isolates of Acanthamoeba. Invest Ophthalmol vis Sci. 2003;44(3):1142–1149. doi: 10.1167/iovs.02-0485. [DOI] [PubMed] [Google Scholar]
- Ledee DR, Iovieno A, Miller D, Mandal N, Diaz M, Fell J, Fini ME, Alfonso EC. Molecular identification of T4 and T5 genotypes in isolates from Acanthamoeba keratitis patients. J Clin Microbiol. 2009;47:1458–1462. doi: 10.1128/JCM.02365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang Z, Qu J, Zhang Y, Sun X. Acanthamoeba keratitis related to contact lens use in a tertiary hospital in China. BMC Ophthalmol. 2019;19(1):202. doi: 10.1186/s12886-019-1210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Morales J, Khan NA, Walochnik J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite. 2015;22:10. doi: 10.1051/parasite/2015010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Morales J, Martínez-Carretero E, Batista N, Álvarez-Marín J, Bahaya Y, Walochnik J, Valladares B. Early diagnosis of amoebic keratitis due to a mixed infection with Acanthamoeba and Hartmannella. Parasitol Res. 2007;102:167–169. doi: 10.1007/s00436-007-0754-x. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Morales J, Martín-Navarro CM, López-Arencibia A, Arnalich-Montiel F, Piñero JE, Valladares B. Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends in Parasitol. 2013;29:4. doi: 10.1016/j.pt.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Morales J, Morcillo-Laiz R, Martín-Navarro CM, López-Vélez R, López-Arencibia A, Arnalich-Montiel F, Maciver SK, Valladares M-C. Acanthamoeba keratitis due to genotype T11 in a rigid gas permeable contact lens wearer in Spain. Cont Lens Anterior Eye. 2011;34(2):83–86. doi: 10.1016/j.clae.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Maghsood AH, Sissons J, Rezaian M, Nolder D, Warhurst D, Khan NA. Acanthamoeba genotype T4 from the UK and Iran and isolation of the T2 genotype from clinical isolates. J Med Microbiol. 2005;54:755–759. doi: 10.1099/jmm.0.45970-0. [DOI] [PubMed] [Google Scholar]
- Marciano-Cabral F, Cabral G. Acanthamoeba as agents of disease in humans. Clin Microbiol Rev. 2003;16:273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Pérez T, Criado-Fornelio A, Martínez J, Blanco MA, Fuentes I, Pérez-Serrano J. Isolation and molecular characterization of Acanthamoeba from patients with keratitis in Spain. Eur J Protistol. 2017;61:244–252. doi: 10.1016/j.ejop.2017.06.009. [DOI] [PubMed] [Google Scholar]
- Maycock NJR, Jayaswal R. Update on Acanthamoeba keratitis. Cornea. 2016;35:713–720. doi: 10.1097/ICO.0000000000000804. [DOI] [PubMed] [Google Scholar]
- Mubareka S, Alfa M, Harding GK, Booton G, Ekins M, VanCaeseele P. Acanthamoeba species keratitis in a soft contact lens wearer molecularly linked to well water. Can J Infect Dis Med Microbiol. 2006;17:2. doi: 10.1155/2006/169625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munguía BC, Molina MO, Gonzáles-Lázaro M, Gonzáles Lezrobles AG, Bonilla P, Martínez-Palomo A. Ultrastructural study of encystation and excystation in Acanthamoeba castellanii. J Eukaryot Microbiol. 2005;52(2):153–158. doi: 10.1111/j.1550-7408.2005.04-3273.x. [DOI] [PubMed] [Google Scholar]
- Nagyová V, Nagy A, Janecek S, Timko J. Morphological, physiological, molecular and phylogenetic characterization of new environmental isolates of Acanthamoeba spp. from the region of Bratislava. Slovakia Biologia. 2010;65:81–91. doi: 10.2478/s11756-009-0217-1. [DOI] [Google Scholar]
- Nagyová V, Nagy A, Timko J. Morphological, physiological and molecular biological characterisation of isolates from first cases of Acanthamoeba keratitis in Slovakia. Parasitol Res. 2010;106(4):861–872. doi: 10.1007/s00436-010-1731-3. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Weisenthal RW, Florakis GJ, Reidy JJ, Gaster RN, Tom D. Penetrating keratoplasty in active Acanthamoeba keratitis. Cornea. 2010;29:1000–1004. doi: 10.1097/ICO.0b013e3181cc79a1. [DOI] [PubMed] [Google Scholar]
- Niyyati M, Lorenzo-Morales J, Rezaie S, Rahimi F, Martín-Navarro CM, Mohebali M, Maghsood AH, Farnia S, Valladares B, Rezaeian M. First report of a mixed infection due to Acanthamoeba genotype T3 and Vahlkampfia in a cosmetic soft contact lens wearer in Iran. Exp Parasitol. 2010;126(1):89–90. doi: 10.1016/j.exppara.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Niyyati M, Lorenzo-Morales J, Rezaie S, Rahimi F, Mohebali M, Maghsood AH, Motevalli-Haghi A, Martín-Navarro CM, Farnia S, Valladares B, Rezaeian M. Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp Parasitol. 2009;121(3):242–245. doi: 10.1016/j.exppara.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Niyyati M, Rezaeian M. Current status of Acanthamoeba in Iran: a narrative review article. Iran J Parasitol. 2015;10(2):157–163. [PMC free article] [PubMed] [Google Scholar]
- Noorjahan NP. Pathogenesis of Acanthamoeba keratitis. Ocul Surf. 2010;8:70–79. doi: 10.1016/s1542-0124(12)70071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes TET, Brazil NT, Fuentefria AM, Rott MB. Acanthamoeba and Fusarium interactions: a possible problem in keratitis. Acta Trop. 2016;157:102–107. doi: 10.1016/j.actatropica.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Nuprasert W, Putaporntip C, Pariyakanok L, Jongwutiwes S. Identification of a novel t17 genotype of Acanthamoeba from environmental isolates and t10 genotype causing keratitis in Thailand. J Clin Microbiol. 2010;48(12):4636–4640. doi: 10.1128/JCM.01090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo BD. Infections caused by free-living amebas. Historical commentaries, taxonomy and nomenclature, protozoology and clinicopathologic features. Rev Chilena Infectol. 2006;23:200–214. doi: 10.4067/S0716-10182006000300002. [DOI] [PubMed] [Google Scholar]
- Omaña-Molina M, Vanzzini-Zago V, Hernandez-Martínez D, Gonzalez-Robles A, Salazar-Villatoro L, Ramirez-Flores E, Oregon-Miranda E, Lorenzo-Morales J, Martinez-Palomo A. Acanthamoeba genotypes T3 and T4 as causative agents of amoebic keratitis in Mexico. Parasitol Res. 2016;115(2):873–878. doi: 10.1007/s00436-015-4821-4. [DOI] [PubMed] [Google Scholar]
- Omaña-Molina M, Vanzzini-Zago V, Hernández-Martínez D, Reyes-Batlle M, Castelan-Ramírez I, Hernández-Olmos P, Salazar-Villatoro L, González-Robles A, Ramírez-Flores E, Servín-Flores C, Flores-Alvarado V, Alcántara-Castro M, Lorenzo-Morales J. Acanthamoeba keratitis in Mexico: report of a clinical case and importance of sensitivity assays for a better outcome. Exp Parasitol. 2019;196:22–27. doi: 10.1016/j.exppara.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Orosz E, Kriskó D, Shi L, Sándor GL, Kiss HJ, Seitz B, Nagy ZZ, Szentmáry N. Clinical course of Acanthamoeba keratitis by genotypes T4 and T8 in Hungary. Acta Microbiol Immunol Hung. 2019;66(3):289–300. doi: 10.1556/030.66.2019.008. [DOI] [PubMed] [Google Scholar]
- Orosz E, Szentmáry KHJ, Farkas A, Kucsera I, Nagy ZZ. First report of Acanthamoeba genotype T8 human keratitis. Acta Microbiol Immunol Hung. 2018;65(1):73–79. doi: 10.1556/030.65.2018.007. [DOI] [PubMed] [Google Scholar]
- Ozkoc S, Tuncay S, Delibas SB, Akisu C, Ozbek Z, Durak I, Walochnik J. Identification of Acanthamoeba genotype T4 and Paravahlkampfia sp. from two clinical samples. J Med Microbiol. 2008;57:392–396. doi: 10.1099/jmm.0.47650-0. [DOI] [PubMed] [Google Scholar]
- Padzik M, Baltaza W, Szaflik JP, Hendiger E, Dybicz M, Chomicz L (2018) Comparison of chlorhexidine disinfectant in vitro effect on environmental and ocular Acanthamoeba strains, the amoebic agents of human keratitis − an emerging sight-threatening corneal disease in Poland. Ann Parasitol 64(3):229–233. 10.17420/ap6403.157 [DOI] [PubMed]
- Padzik M, Starościak B, Szaflik JP, Pietruczuk-Padzik A, Siczek P, Chomicz L (2016) Assessment of in vitro dynamics of pathogenic Acanthamoeba strains originating from contact lens wearers with infectious keratitis. Ann of Parasitol 62(4):331–336. 10.17420/ap6204.69 [DOI] [PubMed]
- Page MA, Mathers WD. Acanthamoeba keratitis: a 12-year experience covering a wide spectrum of presentations diagnoses and outcomes. J Ophthalmol. 2013;2013:670242. doi: 10.1155/2013/670242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Hammersmith K. Contact lens-related microbial keratitis: recent outbreaks. Curr Opin Ophthalmol. 2008;19:302–306. doi: 10.1097/ICU.0b013e3283045e74. [DOI] [PubMed] [Google Scholar]
- Prithiviraj SR, Rajapandian SGK, Gnanam H, Gunasekaran R, Mariappan P, Singh SS, Prajna L. Clinical presentations, genotypic diversity and phylogenetic analysis of Acanthamoeba species causing keratitis. J Med Microbiol. 2020;69(1):87–95. doi: 10.1099/jmm.0.001121. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Yagita K, Kobayashi A, Oikawa Y, Hussein AIA, Matsumura T, Tokoro M. Genetic characterization of clinical Acanthamoeba isolates from Japan using nuclear and mitochondrial small subunit ribosomal RNA. Korean J Parasitol. 2013;51(4):401–411. doi: 10.3347/kjp.2013.51.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Wu X. Evaluation of three different methods to establish animal models of Acanthamoeba keratitis. Yonsei Med J. 2010;51:121–127. doi: 10.3349/ymj.2010.51.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risler A, Coupat-Goutaland B, Pelandakis M. Genotyping and phylogenetic analysis of Acanthamoeba isolates associated with keratitis. Parasitol Res. 2013;112:3807–3816. doi: 10.1007/s00436-013-3572-3. [DOI] [PubMed] [Google Scholar]
- Rocha-Cabrera P, Reyes-Batlle M, Martín-Navarro CM, Dorta-Gorrín A, López-Arencibia A, Sifaoui I, Martínez-Carretero E, Piñero JE, Martín-Barrera F, Valladares B, Lorenzo-Morales J. Detection of Acanthamoeba on the ocular surface in a Spanish population using the Schirmer strip test: pathogenic potential, molecular classification and evaluation of the sensitivity to chlorhexidine and voriconazole of the isolated Acanthamoeba strains. J Med Microbiol. 2015;64(8):849–853. doi: 10.1099/jmm.0.000103. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martín J, Rocha-Cabrera P, Reyes-Batlle M, López-Arencibia A, Sifaoui I, Rizo-Liendo A, Bethencourt-Estrella CJ, Piñero JE, Lorenzo-Morales J. Presence of Acanthamoeba in the ocular surface in a Spanish population of contact lens wearers. Acta Parasitol. 2018;63(2):393–396. doi: 10.1515/ap-2018-0045. [DOI] [PubMed] [Google Scholar]
- Schroeder JM, Booton GC, Hay J, Nisz IA, Seal DV, Markus MB, Fuerst PA, Byers TJ. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of Acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol. 2001;39:1903–1911. doi: 10.1128/JCM.39.5.1903-1911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol. 2004;34:1001–1027. doi: 10.1016/j.ijpara.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Sharifi N, Botero-Kleiven S, Ohman D, Barragan A, Winiecka-Krusnell J. Genotypic characterization of Acanthamoeba spp. causing ocular infections in Swedish patients: identification of the T15 genotype in a case of protracted keratitis. Scand J Infect Dis. 2010;42:781–786. doi: 10.3109/00365548.2010.490563. [DOI] [PubMed] [Google Scholar]
- Sharma R, Jhanji V, Satpathy G, Sharma N, Khokhar S, Agarwal T. Coinfection with Acanthamoeba and Pseudomonas in contact lens Y associated keratitis. Optom vis Sci. 2013;90:2. doi: 10.1097/OPX.0b013e31827f15b4. [DOI] [PubMed] [Google Scholar]
- Sharma S, Pasricha G, Das D, Aggarwal RK. Acanthamoeba keratitis in non-contact lens wearers in India: DNA typing-based validation and a simple detection assay. Arch Ophthalmol. 2004;122(10):1430–1434. doi: 10.1001/archopht.122.10.1430. [DOI] [PubMed] [Google Scholar]
- Shoff M, Joslin CE, Tu EY, Kubatko L, Fuerst PA. Efficacy of contact lens systems against recent clinical and tap water Acanthamoeba isolates. Cornea. 2008;27:713–719. doi: 10.1097/QAI.0b013e31815e7251. [DOI] [PubMed] [Google Scholar]
- Shoff M, Rogerson A, Schatz S, Seal D. Variable responses of Acanthamoeba strains to three multipurpose lens cleaning solutions. Optom vis Sci. 2007;84:202–207. doi: 10.1097/OPX.0b013e3180339f81. [DOI] [PubMed] [Google Scholar]
- Siddiqui R, Dudley R, Khan NA. Acanthamoeba differentiation: a two-faced drama of Dr Jekyll and Mr Hyde. Parasitology. 2012;139:826–834. doi: 10.1017/S0031182012000042. [DOI] [PubMed] [Google Scholar]
- Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012;5:6. doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanakos G, Tzanetou K, Miltsakakis D, Patsoula E, Malamou-Lada E, Vakalis NC. Genotyping of pathogenic Acanthamoebae isolated from clinical samples in Greece-report of a clinical isolate presenting T5 genotype. Parasitol Int. 2006;55(2):147–149. doi: 10.1016/j.parint.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Stothard DR, Schroeder-Diedrich JM, Awwad MH, Gast RJ, Ledee DR, Rodriguez-Zaragoza S, Dean CL, Fuerst PA, Byers TJ. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J Eukaryot Microbiol. 1998;45:45–54. doi: 10.1111/j.1550-7408.1998.tb05068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher EE, Méabed EMH, Abdallah I, Wahed WA. Acanthamoeba keratitis in noncompliant soft contact lenses users: genotyping and risk factors, a study from Cairo. Egypt J Infect Public Health. 2018;11:377–383. doi: 10.1016/j.jiph.2017.09.013. [DOI] [PubMed] [Google Scholar]
- Tananuvat N, Techajongjintana N, Somboon P, Wannasan A. The first Acanthamoeba keratitis case of non-contact lens wearer with HIV infection in Thailand. Korean J Parasitol. 2019;57(5):505–511. doi: 10.3347/kjp.2019.57.5.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfeek GM, Bishara SAH, Sarhan RM, Taher EE, Khayyal AE. Genotyping, physiological and biochemical characterization of potentially pathogenic Acanthamoeba isolated from the environment in Cairo. Egypt Parasitol Res. 2016;115:1871–1881. doi: 10.1007/s00436-016-4927-3. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS. Free-living amebae as opportunistic agents of human disease. J of Neuroparasitol. 2010 doi: 10.4303/jnp/N100802. [DOI] [Google Scholar]
- Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Schuster FL. Opportunistic free-living amebae, part I. Clin Microbiol Newsl. 2008;30:151–158. doi: 10.1016/j.clinmicnews.2008.09.004. [DOI] [Google Scholar]
- Wagner C, Reyes-Batlle M, Ysea MA, Pérez MVG, Rondón CG, Paduani AJN, Pérez AD, López-Arencibia A, Sifaoui I, Galindo MVP, Suárez EP, Martínez-Carretero E, Valladares B, Piñero JE, Lorenzo-Morales J. Genotyping of clinical isolates of Acanthamoeba genus in Venezuela. Acta Parasitol. 2016;61(4):796–801. doi: 10.1515/ap-2016-0110. [DOI] [PubMed] [Google Scholar]
- Walochnik J, Scheikl U, Haller-Schober EM. Twenty years of Acanthamoeba diagnostics in Austria. J Eukaryot Microbiol. 2015;62:3–11. doi: 10.1111/jeu.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowska M, Cisowska A, Myjak P, Marek J, Jurowskaka-Liput J, Jakubaszko J. Acanthamoeba keratitis in contact lens wearers in Poland. Adv Clin Exp Med. 2006;15:553–555. [Google Scholar]
- Wilhelmus KR, Jones DB, Matoba AY, Hamill MB, Pfugfelder SC, Weikert MP. Bilateral Acanthamoeba Keratitis. Am J Ophthalmol. 2008;145:193–197. doi: 10.1016/j.ajo.2007.09.037. [DOI] [PubMed] [Google Scholar]
- Wopereis DB, Bazzo ML, Macedo JP, Casara F, Golfeto L, Venancio E, Oliveira JG, Rott MB, Caumo KS (2020) Free-living amoebae and their relationship to air quality in hospital environments: characterization of Acanthamoeba spp. obtained from air-conditioning systems. Parasitology 147(7):782–790. 10.1017/S0031182020000487 [DOI] [PMC free article] [PubMed]
- Yera H, Zamfir O, Bourcier T, Ancelle T, Batellier L, Dupouy-Camet J, Chaumeil C. Comparison of PCR, microscopic examination and culture for the early diagnosis and characterization of Acanthamoeba isolates from ocular infections. Eur J Clin Microbiol Infect Dis. 2007;26(3):221–224. doi: 10.1007/s10096-007-0268-6. [DOI] [PubMed] [Google Scholar]
- Yera H, Zamfir O, Bourcier T, Viscogliosi E, Noël C, Dupouy-Camet J, Chaumeil C. The genotypic characterisation of Acanthamoeba isolates from human ocular samples. Br J Ophthalmol. 2008;92(8):1139–1141. doi: 10.1136/bjo.2007.132266. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun X, Wang Z, Li R, Luo S, Jin X, Deng S, Chen W. Identification of 18S ribosomal DNA genotype of Acanthamoeba from patients with keratitis in North China. Invest Ophthalmol vis Sci. 2004;45(6):1904–1907. doi: 10.1167/iovs.03-1073. [DOI] [PubMed] [Google Scholar]
- Zhao G, Sun S, Zhao J, Xie L. Genotyping of Acanthamoeba isolates and clinical characteristics of patients with Acanthamoeba keratitis in China. J Med Microbiol. 2010;59:462–466. doi: 10.1099/jmm.0.016667-0. [DOI] [PubMed] [Google Scholar]