Abstract
The coronaviruses (CoVs), including SARS-CoV-2, the agent of the ongoing deadly CoVID-19 pandemic (Coronavirus disease-2019), represent a highly complex and diverse class of RNA viruses with large genomes, complex gene repertoire, and intricate transcriptional and translational mechanisms. The 3′-terminal one-third of the genome encodes four structural proteins, namely spike, envelope, membrane, and nucleocapsid, interspersed with genes for accessory proteins that are largely nonstructural and called ‘open reading frame’ (ORF) proteins with alphanumerical designations, but not in a consistent or sequential order. Here, I report a comparative study of these ORF proteins, mainly encoded in two gene clusters, i.e. between the Spike and the Envelope genes, and between the Membrane and the Nucleocapsid genes. For brevity and focus, a greater emphasis was placed on the first cluster, collectively designated as the ‘orf3 region’ for ease of referral. Overall, an apparently diverse set of ORFs, such as ORF3a, ORF3b, ORF3c, ORF3d, ORF4 and ORF5, but not necessarily numbered in that order on all CoV genomes, were analyzed along with other ORFs. Unexpectedly, the gene order or naming of the ORFs were never fully conserved even within the members of one Genus. These studies also unraveled hitherto unrecognized orf genes in alternative translational frames, encoding potentially novel polypeptides as well as some that are highly similar to known ORFs. Finally, several options of an inclusive and systematic numbering are proposed not only for the orf3 region but also for the other orf genes in the viral genome in an effort to regularize the apparently confusing names and orders. Regardless of the ultimate acceptability of one system over the others, this treatise is hoped to initiate an informed discourse in this area.
Keywords: Coronavirus, Accessory genes, Open reading frame, Phylogeny, RNA genome, Coronaviridae
Abbreviations: CoV, coronavirus; CoVID-19, coronavirus disease-2019; ORF, open reading frame; SARS, severe acute respiratory syndrome; MERS-CoV, (Middle East Respiratory Syndrome CoV).
Graphical abstract
1. Introduction
Coronaviruses (CoVs) are Baltimore Group IV RNA viruses containing single-stranded, positive sense RNA genome ~30 kb in size, relatively large by RNA viral standards but befitting the remarkable complexity of their gene expression (Masters, 2006; Sawicki et al., 2007). Although brought to recent fame by the human pandemic, COVID-19, the coronaviruses infect a large variety of mammals and birds. Less harmful human CoVs, such as HCoV-NL63, have been known as one of the causative agents of common cold. The previous CoV pandemics of 2012 and 2002 were caused respectively by Middle East respiratory syndrome CoV (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV). In continuation of this nomenclature and due to its genetic proximity with SARS-CoV, the novel CoV responsible for the 2019 pandemic has been named SARS-CoV-2. Sequence comparison has indicated that the human CoV strains evolved from enzootic CoVs of other mammals, mostly bats and via carriers such as the palm civet, by acquiring mutations that allowed them to jump species and become virulent human pathogens (Guan et al., 2003; Chinese, 2004; Li, 2013; Cui et al., 2019; Liu et al., 2020; Helmy et al., 2020; Zhou et al., 2020).
Gene sequencing of naturally occurring coronaviruses and studies of their zoonosis have led to the recognition of four genera, namely Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus (Woo et al., 2012; Cui et al., 2019). The Alpha and Beta coronaviruses are of bat origin and infect only mammals, while Gamma and Delta coronaviruses mainly infect birds. The lethal human pathogen, SARS-CoV-2, is a betacoronavirus, believed to have been transmitted from bats (Cui et al., 2019; Helmy et al., 2020; Zhou et al., 2020).
As with essentially all RNA viruses, the CoV RNA genome (schematically shown on the top of Fig. 2) is transcribed by the virally encoded RNA-dependent RNA polymerase (RdRP), but CoV genomes manifest several complex molecular mechanisms that are not found to co-exist in any other Group IV virus, such as initiation of transcription by leader RNA, generation of a nested set of 3՛-coterminal mRNA, synthesis of polyprotein from polycistronic mRNA, followed by proteolytic cleavage, and translation re-initiation by leaky scanning and programmed ribosomal frame-shift (Sawicki et al., 2007; Baranov et al., 2005; Plant et al., 2010). In all coronaviruses, the 3′ two-thirds of the genome include orf1a and orf1b genes (Sawicki et al., 2005; Gao et al., 2020) that contain coding regions for PP1a and PP1b, separated by a stop codon. This region is translated into PP1a or a large polyprotein, PP1ab, the ratio being dictated by the efficiency of ribosomal frameshifting at the stop codon. The polyprotein is processed into ~16 nonstructural proteins (called ‘NSPs') that together forms the RdRP The rest of the genome encodes four structural proteins, namely spike (S, previously ORF2), envelope (E), membrane (M) and nucleocapsid (N). Interestingly, these latter genes are interspersed with ~8 ORFs, mostly between S and E, and between E and M. A few also occur downstream of the N gene and within the N gene in a different reading frame. Overall, the number, location and identity of the ORFs are variable between the various genera and individual viruses (Cui et al., 2019). The function and expression of these ORFs remain largely unknown, and thus, they are currently designated by numbers 3 to 9, such as ORFs 3, 3a, 3b, 3c, 3d, 6, 7a, 7b, 8a, 8b, 9b, 9c, but not always in precise alphanumerical order. For example, the MERS-CoV genome is assigned the gene order -S-3a-4a-4b-5-E-M- while skipping the number 7. Several ORFs have been studied in model coronaviruses, for instance, in SARS-CoV (a Beta CoV), and also in feline CoV (an Alpha CoV) and avian infectious bronchitis virus (IBV) (a Gamma CoV) (Terada et al., 2019; Menachery et al., 2014; Haijema et al., 2004; Hodgson et al., 2006; Liu and Inglis, 1991). Most studies were conducted with recombinant proteins, expressed in cells transfected with plasmid DNA clones; for the majority of ORFs, however, a definitive evidence that they are actually expressed from the viral gene in the infected cell is lacking.
Fig. 2.
Homology-based phylogeny in the ‘orf3 region’ of the three major CoV genera, color coded as in Fig. 1 (Alpha = red; Beta = blue; Gamma = Green). Examples of genus-specific clusters, both interrupted by another Genus, are marked by boxes. Viral name abbreviations are in Table 1, and details are in Section 3.4.
Coronaviruses are sensitive to cellular interferon (IFN), the major antiviral arm of the infected cell (Clementi et al., 2020; Stroher et al., 2004; Sainz Jr. et al., 2004). Use of recombinant proteins and engineered recombinant viruses revealed that many ORFs suppress the IFN induction and the IFN response pathways; viruses with deletions in such genes are indeed attenuated in IFN-proficient cells and in animals but grow unhindered in IFN-deficient cells (Freundt et al., 2009; Spiegel et al., 2005; Kopecky-Bromberg et al., 2007; Shen et al., 2003; McGoldrick et al., 1999; Tung et al., 1992; Yang et al., 2013; Dedeurwaerder et al., 2014; Siu et al., 2014; Matthews et al., 2014; Niemeyer et al., 2013; Tung et al., 1992; Yang et al., 2013; Dedeurwaerder et al., 2014; Matthews et al., 2014; Siu et al., 2014; Niemeyer et al., 2013; Narayanan et al., 2008; Yount et al., 2005). These proteins are, therefore, considered accessory and generally nonstructural, although several SARS-CoV proteins, such as ORF3a, ORF6, ORF7a, and ORF9b, have been shown to be present in mature virions in small amounts, the significance of which remains unknown (Huang et al., 2006; Huang et al., 2007). Evidently, these accessory proteins play an important role in optimal CoV replication and pathogenesis in the host animal by subverting the innate immune mechanisms. This is reminiscent of the IFN-suppressor proteins of nonsegmented negative-strand RNA viruses of the Paramyxoviridae family, which are also nonstructural and not required for virus replication in cultured IFN-deficient cells, but important for optimal virus replication and pathogenesis in animals (Barik, 2013; Dhar and Barik, 2016; Ribaudo and Barik, 2017). Lastly, selected accessory proteins of CoV have been shown to also regulate cell cycle and cell death when expressed recombinantly in uninfected cells; the ORF3a and ORF3b proteins, for instance, induce cell cycle arrest and apoptosis (Padhan et al., 2008; Khan et al., 2006; Law et al., 2005; Yuan et al., 2005; Yuan et al., 2007; Freundt et al., 2010; Yue et al., 2018). Incidentally, some paramyxoviral IFN-suppressor proteins also exhibit IFN-independent functions, such as the two nonstructural proteins of respiratory syncytial virus, which suppresses apoptosis (Bitko et al., 2007).
In view of their cardinal importance in pathology and cellular physiology, the accessory proteins deserve greater attention; it is also important to tease out the relative contribution of the individual proteins in immune suppression and in any other activity. However, the first roadblock to this endeavor is the currently uncertain identity and number of these ORFs and their apparently diverse genomic location and numerical designation in various CoVs, even within the same genus. In this proof of concept study, I have focused on the region bracketed by the S and E genes and explored this diversity in detail. For brevity, I will sometimes refer to this region as the ‘orf3 region’, eponymously for orf3 (sometimes called orf3a), the commonly numbered first orf after the S gene. Analysis was conducted on the two largest coronavirus genera, namely Alpha and Beta, as they also infect mammals, and therefore, hold significance for human disease. Moreover, only a few Gamma and Delta CoVs have been isolated and sequenced, and the deltacoronaviruses were found to be totally devoid of the orf3 region (Supplementary material 1), such that the S gene is immediately followed by E gene (Cui et al., 2019). Finally, two novel coronaviruses, recently isolated from marine mammals, viz. the beluga whale and the bottlenose dolphin, were found to lack orf3, and were tentatively classified as gammacornaviruses on the basis of their full genome sequence (Supplementary material 1) (Mihindukulasuriya et al., 2008; Woo et al., 2014). However, it is the deltacoronaviruses that lack orf3, as indicated above, whereas the gammacoronavirus genomes, at least the ones that have been sequenced, do contain orf3 regions (IBV and turkey-CoV in Fig. X) (Cui et al., 2019). Thus, the marine mammal CoVs may phylogenetically lie somewhere between a Gamma and a Delta virus, which is also supported from studies of protein structure (Barik, 2020). The orf3 region, therefore, may indirectly make important contributions to the CoV genotype. Overall, the work presented here constitutes a critical analysis of the ORF numbering system in CoV with the goal of better referral and understanding, and also offers several alternative nomenclatures.
2. Material and methods
2.1. Sequence retrieval
All full-length and curated coronavirus genome sequences from the RefSeq collection of NCBI were collected using the query word ‘Coronaviridae’. From 66 sequences thus obtained, repeated genomes of multiple field isolates of very similar viruses were removed, such as a large number of bat coronaviruses from many caves in China and multiple submissions of the same viral genome. The final list contained 22 genomes of nonredundant sequences, representing each of the three genotypes that contained the orf3 region (e.g. Alpha, Beta, Gamma) (Table 1 ; Supplementary material 1). Although this screening may have eliminated interesting variants in all genera, the selection was unbiased and encompassed adequate diversity for this initial, proof-of-concept study.
Table 1.
Coronaviruses sequences analyzed in this study.
| Viral genus and name (listed alphabetically) | Abbreviation used | Accession # |
|---|---|---|
| Genus Alphacoronavirus | ||
| Mi-BatCoV HKU8, AFCD77 | HKU8 | EU420139.1 |
| Mi-BatCoV 1B, AFCD307 | AFCD307 | EU420137 |
| Mi-BatCoV 1A, AFCD62 | AFCD62 | NC_010437.1 |
| HCoV-NL63 | NL63 | KF530114.1 |
| Ro-BatCoV HKU10 | HKU10 | NC_018871.1 |
| Porcine epidemic diarrhea virus | PEDV | AF353511.1 |
| Rhinolophus bat CoV HKU2 | HKU2 | EF203067.1 |
| Porcine respiratory CoV ISU-1 | PRCV | DQ811787.1 |
| Transmissible gastroenteritis virus | TGEV | NC_038861.1 |
| Feline CoV 1Je | FCoV | KU215423.1 |
| BatCoV CDPHE15/USA/2006 | CDPHE | NC_022103.1 |
| Sc BatCoV 512/2005 | 512 | NC_009657.1 |
| Genus Betacoronavirus | ||
| Bat CoV 133/2005 | 133 | DQ648794.1 |
| Bat CoV HKU4–1 | HKU4 | BEF065505.1 |
| Middle Eastern respiratory syndrome CoV | MERS | NC_019843.3 |
| Bat SARSr-CoV HKU3–1 | HKU3 | DQ022305.2 |
| Bat SARS CoV Rp3/2004 | Rp3 | DQ071615.1 |
| SARS CoV Tor2 | Tor2 | AY274119.3 |
| Bat SARS-like coronavirus WIV16 | WIV16 | KT444582.1 |
| SARS-CoV-2 | CoV-2 | NC_045512 |
| Bat CoV HKU9–1 | HKU9 | NC_009021.1 |
| Genus Gammacoronavirus | ||
| Turkey (Avian) coronavirus | TurCoV | NC_010800.1 |
| Avian infectious bronchitis virus | IBV | AJ311362.1 |
CoV = Coronavirus, SARS = Severe acute respiratory syndrome. The accession numbers are for the NCBI GenBank. The generally accepted abbreviations of virus names are listed here, but some names were shortened further in specific Figures for space saving. Note that the same virus is sometimes referred by different names in the literature, such as HKU8 as well as AFCD77.
The sequences were visually examined for the ‘orf3 region’ between the S and the E genes, and the already-annotated ORF names were noted (Supplementary material 1).
2.2. Sequence analysis of ORFs
The conceptually translated ‘orf3 region’ sequence of the select group of viruses was analyzed for coding sequences in three translation frames and only in the forward direction (mRNA-sense, genome 5′ to 3′), using the web site https://web.expasy.org/translate/. The results included both annotated and new ORFs, as listed (Supplemental material 1). The ORF sequences were subjected to multiple alignment using Clustal Omega and default parameters. The similarities were converted to PhyML format and analyzed and plotted by Bayesian Information Criterion (BIC) using PhyMol 3.0 (available on GitHib) (Lefort et al., 2017). Also known as Schwarz Information Criterion, BIC is based in part on the maximum likelihood function, incorporating probabilistic values for phylogenetic branches.
3. Results and discussion
3.1. Prototype coronaviruses analyzed for the orf3 region
For the comparative analysis of Alphacoronavirus and Betacoronavirus orf3 regions, prototype viruses were chosen, representing diverse viral and host species as well as those important in public and animal health, such as SARS CoV and porcine CoV. These viruses and their abbreviated names used in this paper are listed (Table 1).
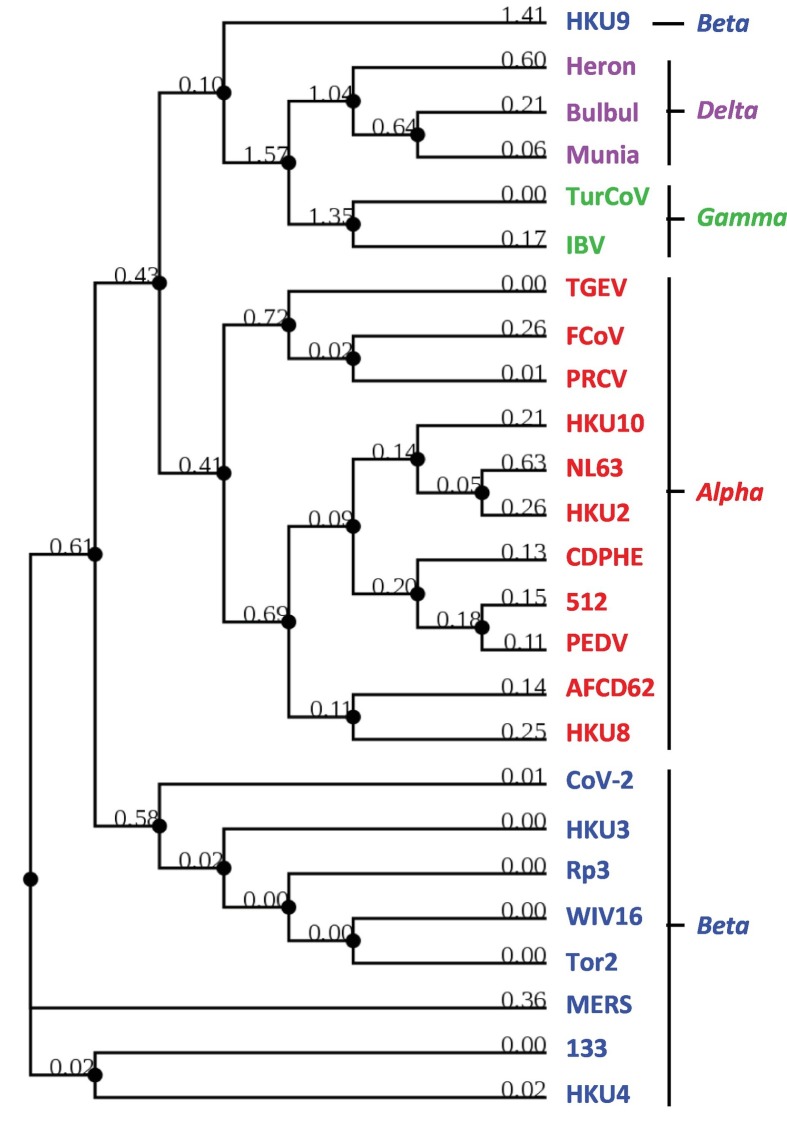
3.2. Phylogeny match of coronaviruses to the envelope protein sequences
As mentioned earlier, the CoVs are classified into four well-established genera, based on the overall sequence of the genome and those of the highly conserved genes, such as the orf1a and orf1b duo, which generates the viral RdRP. To study the sequence similarity among the ORF family proteins, I needed a frame of reference, but the orf1a/b genes were far away from the orf3 region, as the gene arrangement at this end of the CoV genome is 3′-1ab-S-orf3 region-E--. The neighboring E protein was, therefore, chosen and its sequences compared by multiple sequence alignment (Fig. 1 ). The resulting tree in fact neatly matched the Genera, such that all members of a given Genus were clustered together, with the minor exception of the bat isolate HKU9 (a Beta coronavirus) that was closer to Delta and Gamma coronaviruses. Thus, the sequence of the structural protein, E, followed viral Genus, confirming and extending a similar observation with the 1a/1b polyprotein (Cui et al., 2019). Upon establishing the E protein as benchmark, I was in a position to test whether or not the nonstructural ORF proteins also follow a similar Genus-specific pattern.
Fig. 1.
Sequence homology among E proteins from selected coronaviruses, confirming that the E sequences correspond to the Genera, which are color-coded for easy viewing (Alpha = red; Beta = blue; Gamma = Green; Delta = pink). The results match with the phylogeny based on CoV RdRP (Cui et al., 1994). Virus names and abbreviations are listed in Table 1 (Section 3.2). Alignment was performed as described in Materials and Methods, and the branch lengths indicated in the rooted guide tree. For consistency, these same viral strains were used as reference in all analyses in this study, using the same color codes.
3.3. Comprehensive accounting of the ORFs
First, all known orfs in one of the orf clusters in the CoV genomes, namely those in the area I named the ‘orf3 region’ (between S and E genes), were analyzed. To this end, all annotated ORFs, such as 3, 3a, 3b etc. were collected from the NCBI genome accession sites (Table 1), and were confirmed by manual conceptual translation. The predicted polypeptides ranged in length from 70 aa (FCoV-3a) to 244 aa (TGEV-3b). It is to be noted that several of these reported ORFs are in different reading frames. Additionally, all previously unreported ORFs were also collected and tentatively named ORFX; when multiple new ones were found in the same genome, they were named ORFX1, ORFX2 etc. (Supplementary material 2). In this exploratory first study I have adopted an arbitrary length of >55 aa as the cut-off for ORFX, simply to reduce the pool. However, future studies can explore many smaller ORFs, since they may encode interacting functional domains and motifs. As we will also see later, a few X proteins, although not recognized previously, show homology with known ORFs and/or with one another.
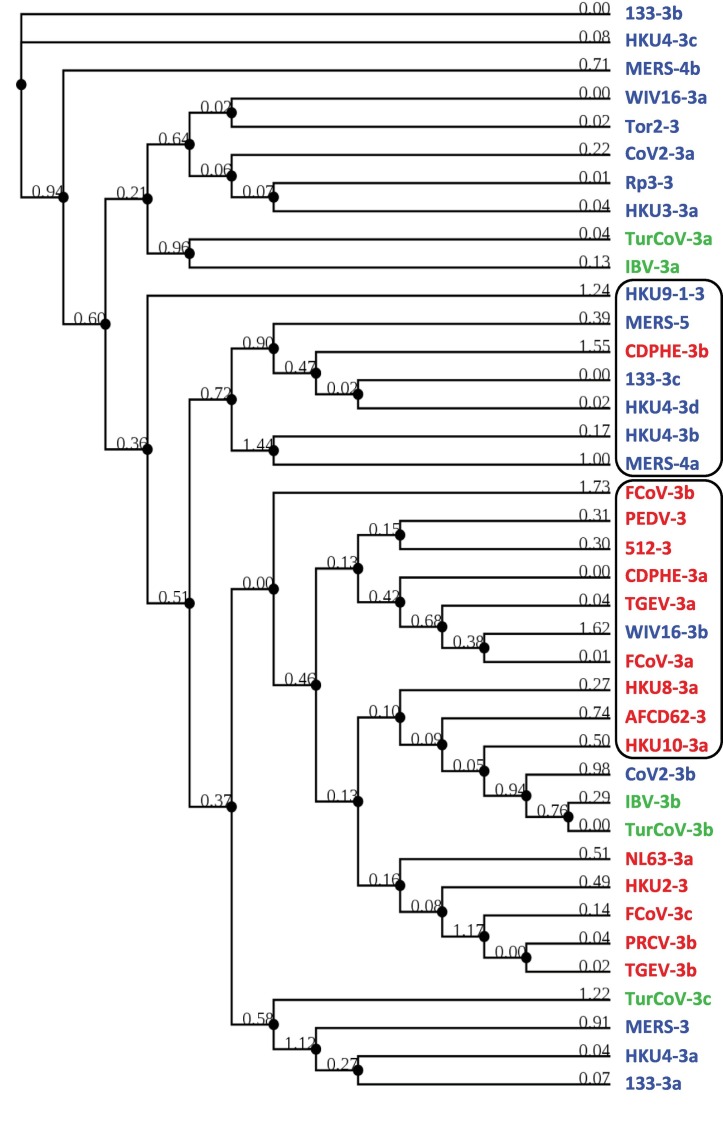
3.4. Sequence relationships among the orf3 region proteins
To determine the relationship between the apparently disparate numerical designations of these ORFs in various CoVs, multiple alignment of all known and new ORFs was performed using Clustal Omega and Bayesian Information Criterion as described in Section 2.2, and the result is shown (Fig. 2 ). When the sequences were identified with the viral genera, it revealed several small similarity clusters, but the overall homology did not follow the genera, in sharp contrast to what we saw with the structural proteins (Fig. 1). The ORF names were also scattered; for example, ORF3b of 133 was similar to ORF3c of HKU4, as were ORF3a of WIV16 and ORF3 of Tor2.
Nonetheless, closer scrutiny revealed the existence of several clusters, each of which was viral genus-specific, i.e., Alpha (red) or Beta (blue), but not both (Fig. 2). However, even this had exceptions, in which an otherwise Genus-specific cluster was interrupted by a homolog from a different Genus. Two such examples are marked (Fig. 2), where CDPHE-3b disrupted a Beta cluster, and WIV16-3b disrupted an Alpha cluster. Regardless, many clusters had mixed ORF numbers; for example, one contained ORFs 3, 5, 3b, 3d, and 4a. This confirmed our earlier impression that even the homologous sequences in different CoVs were inconsistently numbered. Because of their potentially novel nature, the ORFX's were analyzed separately, as described later.
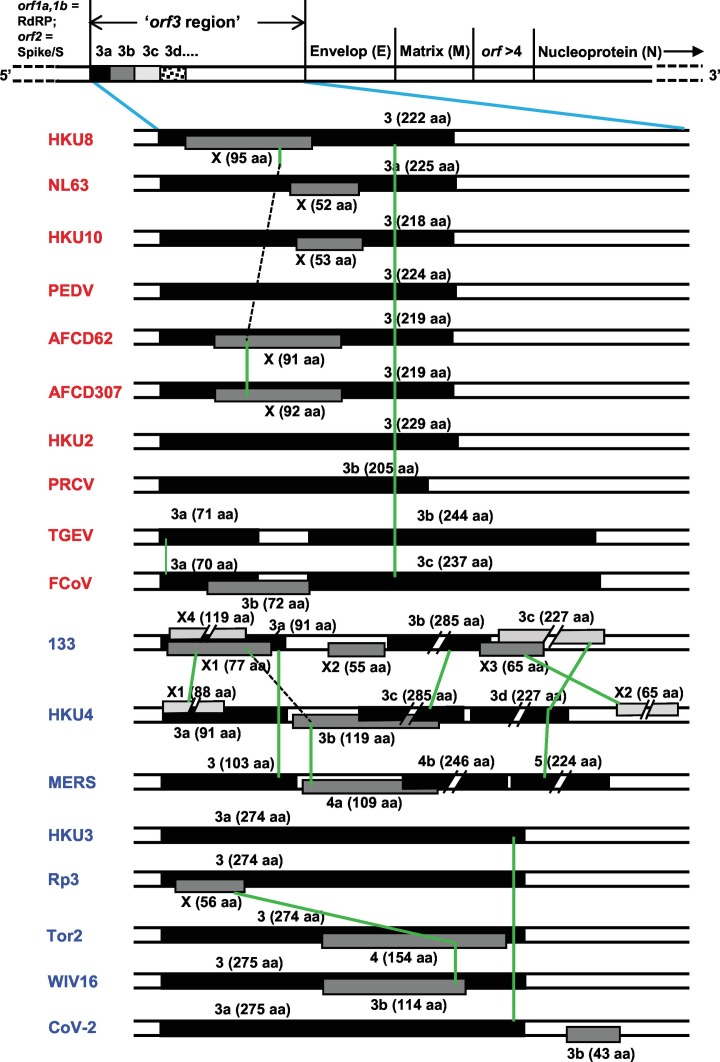
3.5. Lack of shared synteny of orthologous ORFs among CoV genomes
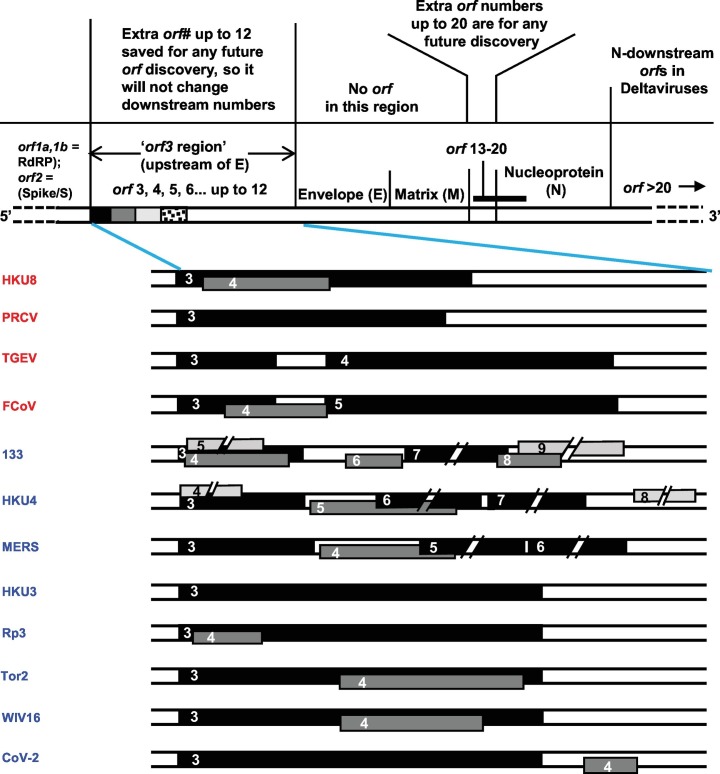
Location of orthologous genes are often found conserved between two sets of chromosomes; this phenomenon and other locational similarities are often referred to as ‘synteny’ or ‘shared synteny’ (Zhao and Schranz, 2017; Adato et al., 2015). Reciprocally, presence of synteny tends to confirm similarity of genetic loci and horizontal gene transfer. With this in mind, an analysis of the ‘orf3 region’ gene locations was undertaken, using selected representative viruses. For ease of visualization, simplified schematic drawings of this region were lined up and the homologs were connected. The results (Fig. 3 ) graphically reveal a clear lack of synteny in that there are very few ORFs of any given cluster that are in the same vertical line, most being at an angle with each other, i.e., shifted in location.
Fig. 3.
Comparative ORF locations in the ‘orf3 region’ of two major CoV genomes, depicting the currently used names. Virus names are as in the previous Figures. The genetic map of a CoV is schematically shown on top, not drawn to scale. When multiple translational frames were used, they were indicated by different thickness and shade; the main frame (frame 0), considered as the one used by the most 5′-proximal (left side in the diagram) orf in the genome (usually ORF3/3a; 3b, etc.) in black color, and the −1 and − 2 frames are progressively lighter. The ORF lengths were drawn approximately to scale, except when a long sequence was truncated in the middle in order to fit it in the available space. For these truncated ORFs, only the left terminus, and not the right, is properly positioned. To indicate the actual lengths, the amino acid (aa) numbers encoded in all ORFs are shown in parenthesis. ORF overlaps are indicated by placing them in different tracks (over one another); however, due to space constraints, specific tracks could not be assigned to each translational frame. Orthologous ORFs are connected by green lines and any dissimilar ORFs between them were skipped and indicated by broken line segments in black color. Further details are in Section 3.5. Note that all viruses in this Figure are also contained in Fig. 2, with the sole exception of AFCD307 as it is essentially identical to AFCD62.
Among the multiple viral examples shown, a few generalizations can be noted here. ORF3, −3a, −3b, and -3c – all >200 aa long – are often essentially the same gene, i.e. orthologous; however, the ORF3a of TGEV and FCoV, respectively 71 and 70 aa long and highly similar to each other (69% identical residues), are very different from all other 3-series ORFs. Likewise, ORF3b in both FCoV and BatCoV HKU4, respectively 72 and 119 aa long, are entirely different in sequence. In fact, the BatCoV HKU4 ORF3b is similar to the newly mined ORFX1 of BatCoV133, another bat virus, and to ORF4a of MERS. In most viruses, the ORF3 region numbering did not exceed the number 4, with a few exceptions, namely, the ORF5 MERS and ORF6 in Delta-CoV HKU11 (not shown). MERS ORF5 is actually orthologous to ORF3d of BatCoV HKU4 and ORF3a of BatCov133.
In continuation of these studies, further attention was paid to the possible identity of the newly mined ‘ORFX’ group (Supplementary material 2). Their genomic locations were shown in representative viruses (Fig. 3), which revealed diversity as well as distinction from the previously numbered genes. In exploring their identity, two ORFX polypeptides were found to be close homologs of known ORFs (Fig. 4 ). Homology search by BLAST readily revealed that 133-X4 is essentially identical to what has been variously named ORF 3b, 4, 4a, and gp4 in different members of the HKU4 CoV family, one of which is shown here (Fig. 4A). Likewise, Rp3-X was found to be orthologous to several ORF3b and also to ORF4 in two civet (a small mammal) SARS-CoVs, viz. 010 and 007/2004; however, it is much smaller due to a shorter C-terminus (Fig. 4B).
Fig. 4.
Identity of two unnamed CoV ORFs, viz. 133-X4 (A) and Rp3-X (B). The predicted amino acid sequences were used as query in BLAST, and representative orthologous ORFs are shown in alignment. Note that the homologs are annotated with various ORF numbers in various viruses even when they are highly related, such as the 133 and HKU4 family viruses, both betacoronaviruses from bats (Table 1). The NCBI accession number of Civet010 ORF4 is AAU04651.1.
All other ORFX showed no significant sequence similarity with known ORFs, but showed some similarity with another X ORF (Fig. 5 ). HKU8-X and HKU10-X were very different from the others but were relatively close to each other. They also revealed no orthologs in a broader BLAST search, outside of the Coronaviridae. Lack of orthologs suggests that such ORFs are unique to specific viruses, and therefore, may encode exclusive functions; alternatively, they are false positives, non-functional ORFs, and as such, not conserved.
Fig. 5.
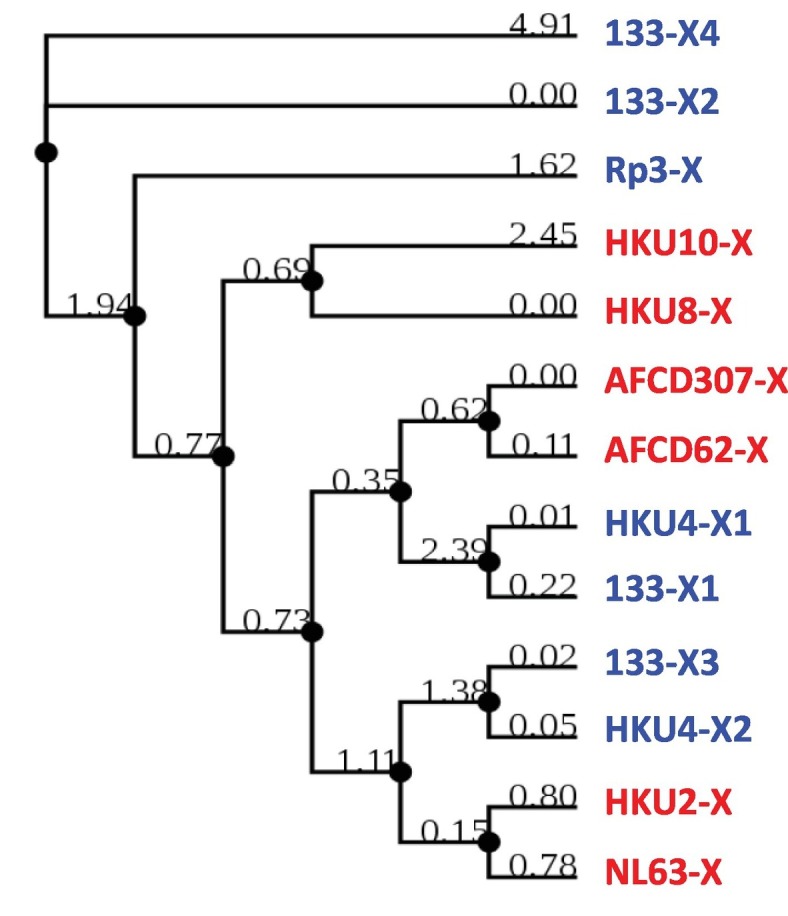
Phylogeny of newly mined CoV ORFs (ORFX#). The conceptually translated sequences of hitherto uncharacterized ORFs in the set of CoVs were subjected to multiple sequence alignment and the homology tree drawn as described in Materials and Methods. The two Genera are color-coded as before (Alpha = red; Beta = blue). Note that the similarity branches of the total ORFX sequences do not cluster by viral Genus, but there is a trend of local clustering within each genus.
Lastly, all ORFX, with only two exceptions, namely ORFX2 of BatCoV 133 and BatCoV HKU4 (55 aa and 65 aa, respectively) are located internal to other local ORFs, as clearly seen within the 3-series, i.e. 3, 3a, 3b, 3c, but translated in different reading frames (indicated by distinctive shades in Fig. 3). The most extensively overlapping ones include HKU4-X1 (88 aa), CoV 133-X3 (65 aa), and the relatively large X4 (119 aa) in 133, which together overlap with 3a, 3b and 3c. The ORFX of Rp3 is completely internal to ORF3. Additionally, the two new ORFs, X1 and X4, of 133 overlap each other in separate translational frames (Fig. 3). Other notable overlaps are exemplified by a total inclusion of small X ORFs inside ORF3/3a in several Alpha viruses, and nearly complete overlap of X3 with 3b and 3c in 133, complete inclusion of 4 of SARS-CoV Tor2 and 3b of Bat SARS-CoV WIV16 within their respective orf3 genes. In view of such widespread gene overlaps, the few coronaviruses with apparently a single ORF in the ORF3 region stood out, such as PEDV and HKU2 (ORF3 only), PRCV (ORF3b only), and HKU3 (ORF3a only) (Fig. 3).
The enormous variety of gene repertoire and gene order in coronaviruses, as revealed here (Fig. 2, Fig. 3), is extraordinary among viruses, even considering the high mutation rate of RNA genomes (Domingo and Holland, 1997). In nonsegmented negative-strand RNA viruses, for example, the gene homologs and their relative locations are largely conserved within each family, such as the gene order N-P-M-G-L in the large Rhabdoviridae family (Banerjee and Barik, 1992). Similarly, in Paramyxoviridae family, the gene order in majority of viruses is N-V/P-M-F-SH-HN-L, although minor variations, such as presence or absence of the SH gene, which is nonessential in cell culture, can be seen (Banerjee et al., 1991). In these viruses, the V/P gene generates two different but overlapping proteins through transcriptional insertion of non-templated G in the mRNA by a process known as RNA editing, leading to translational frame-shift. In the two highly related respiroviruses, hRSV (human respiratory syncytial virus) and PVM (pneumonia virus of mice), although they infect different animals, the gene order is conserved as NS1-NS2-N-P-M-SH-G-F-M2-L, of which NS1 and NS2 code for nonstructural proteins (Barik, 2013; Dhar and Barik, 2016). The closely related Pneumovirus genus, exemplified by the human metapneumovirus (hMPV), contains all the homologs of the structural protein genes, but interestingly, the SH-G gene cassette (underlined) has shifted position towards the 5′ end of the genome, resulting in the gene order N-P-M-F-M2-SH-G-L (Hamelin and Boivin, 2005). In coronaviruses, even within the same genus, such shifts and reorganizations are much more extensive as well as variable (Fig. 2, Fig. 3). Moreover, there is ample diversity even in the use of translational frames among the homologous ORFs (Fig. 2, Fig. 3).
Part of the diversity of coronaviruses is certainly due to the high propensity of recombination between their RNA genomes, potentially involving a variety of molecular mechanisms (Nagy and Simon, 1997; Rowe et al., 1997). In the SARS family of bat coronaviruses, the crowding of bats in the same cave system may also promote rapid exchange of viruses of different strains among the bats, offering a fertile ground for recombination and creating new varieties, which may underlie the apparent interruptions of one genus by another in sequence-based phylogeny tree, such as in Fig. 2.
3.6. Current status and future plans for CoV ORF nomenclature
It is clear from all the aforesaid that the lack of a universal convention has spawned nearly random naming of the viral ORFs by different laboratories when a new CoV is isolated and sequenced, causing significant confusion to the researchers and the general readership alike. The major difficulties in creating a rational convention for CoV have been: the variable gene locations from one virus to another, multiple overlapping ORFs accessing different translational frames in the same sequence, and scarcity of knowledge about the function of the ORFs. Even when functional studies of an ORF were conducted, seemingly multiple roles were unraveled, such as suppression of innate immunity as well as regulation of cell cycle and apoptosis (Section 1), thus adding to the obstacles of creating a universally acceptable function-based name.
Nevertheless, we can explore a few naming strategies, at least to initiate a discourse. First of all, naming a strictly hypothetical ORF that is not actually expressed would be futile, because it would add a nonexistent ORF to the family and may affect all ORF numbers. The easiest experimental approach would be to generate synthetic peptide antibodies against the predicted sequence and use it to probe the infected cell lysate in immunoblot. Discovering the function of all ORFs – even their major functions –will require long-term research and thus, function-based naming is presently not possible. I propose the following strategies in the interim:
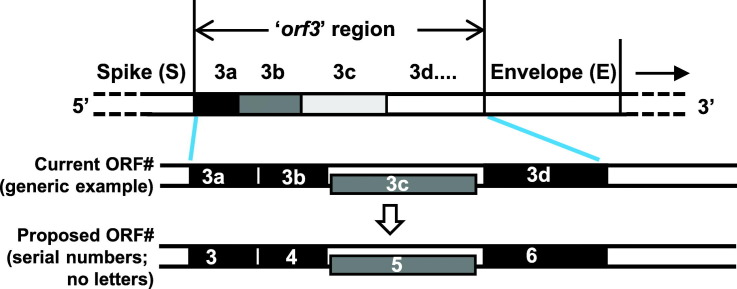
3.6.1. Serial numbering
This is a straightforward strategy, and perhaps the most feasible. In it, the ORFs are named numerically (or alphanumerically, if necessary), simply going from the 5′ end of the CoV genome (Fig. 6 ). Increasing numbers are assigned to the next ORFs as they are encountered, regardless of their 3′ end or overlap with other ORFs; translational frames are also ignored. A few examples are shown in Fig. 6.
Fig. 6.
Proposed new nomenclature for the orf3 region. This is drawn in the same format as Fig. 3 for easy comparison between the two, and shows the proposed new numbering of the ORFs in a subset of genomes for illustration purposes. As in Fig. 3, a schematic of the ORF locations on a generic CoV genome is shown on top, with brief rationales above it, and detailed in Section 3.6. The extra ORF numbers, although not currently required, have been added for contingency, such that if new ORFs are discovered and added to any of these regions in the future, they will not affect the downstream ORF numbers. Some current ORFs between M and N actually overlap with the N sequence, which is also indicated for ORF 13–20 in this region.
A comparison with Fig. 3 will immediately reveal that the new system is much simpler, and that the X's are also numbered. In the schematic example (Fig. 6), I have added a few extra numbers as placeholders for new ORFXs, such as smaller proteins, which may be characterized in the future. Nevertheless, this strategy also has its limitations, the most notable of which is that orthologous ORFs in different viruses may receive different numbers because of their different relative location. For example, ORF3b of 133 and ORF3c of HKU4 are orthologs (note the green line connecting them in Fig. 3), but they will be renamed ORF7 and ORF6, respectively. However, the current system is no better in this respect, since ORF3b and ORF3c are also different names. In many more cases, the newer system actually assigns the same number to similar ORFs, bringing harmony. For example, the first ORFs in five viruses, viz. SARS-CoV-2, BatCoV HKU3, Bat SARS-CoV Rp3, SARS-CoV Tor2, and Bat SARS-CoV WIV16, which are all homologous, currently have different numbers, i.e. 3 and 3a (Fig. 3); in the new system they are all ORF3 (Fig. 6). Similarly, the second ORFs in the last three viruses, which are also homologous, are currently named X, 4, and 3b, respectively; in the new system, they are all ORF4.
3.6.2. Include the translation frame into the nomenclature
This is not helpful because of the frequent use of the same frame by multiple diverse ORFs, as well as different translational frames by orthologous ORFs (Fig. 3). For instance, in TGEV and FCoV, frame 0 houses ORF3, ORF4 and ORF3, ORF5, respectively (Fig. 3). Similarly, ORF3c of BatCoV 133 is translated in frame −2, whereas the homologous ORF3d of another bat virus, namely BatCoV HKU4, is in frame 0.
3.6.3. Maintain the status quo
That has been the approach so far, and therefore, not acceptable for (Section 1).
On a peripheral note, a new, institutionalized nomenclature system will require input from an international consortium of coronavirus experts analyzing all available sequences, and a collective will to reach a consensus. Perhaps the complexity can be made more manageable by dividing the homologs to each genus and adding the genus prefix to the ORF, such as αORF3, βORF3. Regardless, I would submit that the need is urgent since it is common knowledge that old names take root in the explosively growing literature, making it increasingly harder to alter them. However, history has proven that it is possible, since substantial changes in convention have been made quite often in many biological areas. A large-scale example is the renaming of Ser/Thr protein phosphatases (PP) to phosphoprotein phosphatase (PPP), so that the prototype members PP1 and PP2C became Ppp1c and Ppm1c, respectively (Bheri et al., 2020).
3.7. Conclusions
Close examination of the ORFs of the extant coronavirus genomes readily revealed an erratic numbering pattern that differed from virus to virus. A serial numbering plan is proposed based on sequence similarity, which will also include currently unrecognized ORFs, thus allowing a better view of the genetic profile and evolutionary landscape of this highly variable RNA virus and facilitating the analysis of viral behavior, vaccine development and optimal drug targets.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author statement
Sailen Barik, the sole author, performed all aspects of the paper.
Declaration of Competing Interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104858.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- Adato O., Ninyo N., Gophna U., Snir S. Detecting horizontal gene transfer between closely related taxa. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- Banerjee A.K., Barik S., De B.P. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol. Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. Respiratory syncytial virus mechanisms to interfere with type 1 interferons. Curr. Top. Microbiol. Immunol. 2013;372:173–191. doi: 10.1007/978-3-642-38919-1_9. [DOI] [PubMed] [Google Scholar]
- Barik S. Genus-specific pattern of intrinsically disordered central regions in the nucleocapsid protein of coronaviruses. Comput. Struct. Biotechnol. J. 2020;18:1884–1890. doi: 10.1016/j.csbj.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheri M., Mahiwal S., Sanyal S.K., Pandey G.K. Plant protein phosphatases: What do we know about their mechanism of action? FEBS J. 2020 doi: 10.1111/febs.15454. Jun 15. [DOI] [PubMed] [Google Scholar]
- Bitko V., Shulyayeva O., Mazumder B., Musiyenko A., Ramaswamy M., Look D.C., Barik S. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-kappaB-dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 2007;81:1786–1795. doi: 10.1128/JVI.01420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Clementi N., Ferrarese R., Criscuolo E., Diotti R.A., Castelli M., Scagnolari C., Burioni R., Antonelli G., Clementi M., Mancini N. Interferon-β 1a inhibits SARS-CoV-2 in vitro when administered after virus infection. J. Infect. Dis. 2020;225:722–725. doi: 10.1093/infdis/jiaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder A., Olyslaegers D.A., Desmarets L.M., Roukaerts I.D., Theuns S., Nauwynck H.J. ORF7-encoded accessory protein 7a of feline infectious peritonitis virus as a counteragent against IFN-α-induced antiviral response. J. Gen. Virol. 2014;95:393–402. doi: 10.1099/vir.0.058743-0. [DOI] [PubMed] [Google Scholar]
- Dhar J., Barik S. Unique nonstructural proteins of pneumonia virus of mice (PVM) promote degradation of interferon (IFN) pathway components and IFN-stimulated gene proteins. Sci. Rep. 2016;6:38139. doi: 10.1038/srep38139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Park E., Lenardo M.J., Xu X.-N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R., Lippincott-Schwartz J., Zaki S.R., Xu X.-N., Baric R.S., Subbarao K., Lenardo M.J. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84:1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/jvi.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin M.-È., Boivin G. Human metapneumovirus: a ubiquitous and long-standing respiratory pathogen. Pediatr. Infect. Dis. J. 2005;24(11 Suppl):S203–S207. doi: 10.1097/01.inf.0000188158.27840.7c. [DOI] [PubMed] [Google Scholar]
- Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9 doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Narayanan K., Ito N., Peters C., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 2006;80:210–217. doi: 10.1128/JVI.80.1.210-217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 2007;81:5423–5426. doi: 10.1128/JVI.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Fielding B.C., Tan T.H., Chou C.-F., Shen S., Lim S.G., Hong W., Tan Y.-J. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20–27. doi: 10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M., Tsui S.K.W. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Lefort V., Longueville J.-E., Gascuel O. SMS: smart model selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013;100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Kuo R.L., Shih S.R. COVID-19: the first documented coronavirus pandemic in history. Biom. J. 2020;S2319-4170:30044–30045. doi: 10.1016/j.bj.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick A., Lowings J.P., Paton D.J. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 1999;144:763–770. doi: 10.1007/s007050050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2'-O-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya K.A., Wu G., St Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T., Barchet W., Weber F., Drosten C., Müller M.A. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K., Minakshi R., Towheed M.A.B., Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Rakauskaite R., Taylor D.R., Dinman J.D. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaudo M., Barik S. The nonstructural proteins of Pneumoviruses are remarkably distinct in substrate diversity and specificity. Virol. J. 2017;14:215. doi: 10.1186/s12985-017-0881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe C.L., Fleming J.O., Nathan M.J., Sgro J.Y., Palmenberg A.C., Baker S.C. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol. 1997;71:6183–6190. doi: 10.1128/JVI.71.8.6183-6190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr., Mossel E.C., Peters C.J., Garry R.F. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1(4) doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Wen Z., Liu D. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311:16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Yeung M.L., Kok K.-H., Yuen K.-S., Kew C., Lui P.-Y., Chan C.-P., Tse H., Woo P.C.Y., Yuen K.-Y., Jin D.-Y. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., Weber F. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroher U., DiCaro A., Li Y., Strong J.E., Aoki F., Plummer F., Jones S.M., Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon-alpha. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Kuroda Y., Morikawa S., Matsuura Y., Maeda K., Kamitani W. Establishment of a virulent full-length cDNA clone for type I feline coronavirus strain C3663. J. Virol. 2019;93:e01208–e01219. doi: 10.1128/JVI.01208-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung F.Y., Abraham S., Sethna M., Hung S.-L., Sethna P., Hogue B.G., Brian D.A. The 9-kDa hydrophobic protein encoded at the 3 end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and aviancoronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K., Lam C.S., Tsang A.K., Hui S.W., Fan R.Y., Martelli P., Yuen K.Y. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J. Virol. 2014;88:1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yao Z., Wu J., Zhou Y., Shan Y., Dong B., Zhao Z., Hua P., Chen J., Cong Y. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am. J. Respir. Cell Mol. Biol. 2007;37:9–19. doi: 10.1165/rcmb.2005-0345RC. [DOI] [PubMed] [Google Scholar]
- Yue Y., Nabar N.R., Shi C.-S., Kamenyeva O., Xiao X., Hwang I.-Y., Wang M., Kehrl J.H. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Schranz M.E. Network approaches for plant phylogenomic synteny analysis. Curr. Opin. Plant Biol. 2017;36:129–134. doi: 10.1016/j.pbi.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2