Abstract
The chromatin-based DNA damage response pathway is tightly orchestrated by histone post-translational modifications, including histone H2A ubiquitination. Ubiquitination plays an integral role in regulating cellular processes including DNA damage signaling and repair. The ubiquitin E3 ligase RNF168 is essential in assembling a cohort of DNA repair proteins at the damaged chromatin via its enzymatic activity. RNF168 ubiquitinates histone H2A(X) at the N-terminus and generates a specific docking scaffold for ubiquitin binding motif-containing proteins. The regulation of RNF168 at damaged chromatin and the mechanistic implication in the recruitment of DNA repair proteins to the damaged sites remains an area of active investigation. Here, we review the function and regulation of RNF168 in the context of ubiquitin-mediated DNA damage signaling and repair. We will also discuss the unanswered questions that require further investigation and how understanding RNF168 targeting specificity could benefit the therapeutic development for cancer treatment.
Keywords: RIDDLIN, RIDDLE syndrome, DNA repair, DNA double strand break, ubiquitin
Introduction
Our genome is constantly challenged by exogenous genotoxins and endogenous reactive metabolites which result in DNA damage. Unrepaired DNA damage ultimately leads to DNA double strand breaks (DSBs), the most deleterious form of DNA damage in cells, which can lead to genomic instability and cell death if not repaired. Cells have evolved a surveillance system, the DNA damage response (DDR) pathway, to sense, signal, and repair DNA breaks [1, 2]. In particular, DSBs occur in the context of chromatin, which is a bona fide DDR substrate [3]. DSBs are repaired primarily by homologous recombination (HR), non-homologous end joining (NHEJ), alternative end joining (alt-EJ), or single strand annealing (SSA) pathways. Mechanisms regulating DSB repair pathway choice has been extensively discussed by several reviews [4–6].
DSBs elicit an array of post-translational modifications on histones, and DNA repair proteins which promote or regulate DNA damage signaling, recruitment of DNA repair proteins to the DSBs, and DNA repair metabolism [7–11]. The interwoven relationship of these processes dictates the repair choice outcome spatiotemporally via the DDR pathway [9]. In particular, histone ubiquitination participates in the complex interplay of modifications on chromatin to regulate biological processes, including the DDR pathway by amplification of damage signals and downstream recruitment and retention of repair proteins at damaged chromatin [12, 13]. In this review, we will focus primarily on the RING finger protein 168 (RNF168, also known as RIDDLIN), an E3 ligase that plays an essential role in regulating DDR signaling and repair by catalyzing the ubiquitination of histones and other DDR proteins.
Ubiquitination (ubiquitylation or ubiquitinylation) is a process by which an E1 (ubiquitin activating enzyme), E2 (ubiquitin conjugating enzyme) and E3 (ubiquitin ligase) covalently conjugate ubiquitin to the amino group of a protein’s N-terminus or to lysine residues within the substrates [14]. Ubiquitination of proteins serves as a signal regulating protein degradation, cellular localization of proteins, activation and inactivation of enzymes, signal transduction and protein-protein interaction. Ubiquitin can be conjugated singularly onto the lysine residue (monoubiquitination) or as a polyubiquitin chain. [14, 15]. Polyubiquitination chains are formed by linking the glycine residue of a ubiquitin moiety to a lysine residue (K6, K11, K27, K29, K33, K48 and K63) or the N-terminal methionine residue (M1) of a ubiquitin that is conjugated to the lysine residue of the substrate [15, 16].
RNF168, a 571-amino acid nuclear protein, is a central player in the DDR pathway [17–19]. RNF168 was discovered with its association with RIDDLE (acronym of the major features; Radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties) syndrome in 2009 [18] and understanding of RNF168 regulation and its biological functions has advanced rapidly in the last decade. Herein, we review the current literature on RNF168’s molecular functions and cellular biology with particular focus on its recently characterized substrate recognition mechanism and its potential implications in orchestrating DNA DSB repair.
1. RNF168/RIDDLIN and RIDDLE syndrome
RIDDLE syndrome was first reported in 2007 as a rare genetic disease [20]. RIDDLE syndrome patients exhibit differential clinical phenotypic spectrum including reduced immunoglobulin, ataxia, microcephaly, learning difficulties, and respiratory failure. Cells isolated from RIDDLE syndrome patients display impairment of tumor suppressor p53 binding protein 1 (53BP1) recruitment to DSBs, resulting in ionizing radiation hypersensitivity and cell cycle checkpoint defects [20]. Patients with RIDDLE syndrome show pathological similarities to the ataxia-telangiectasia (AT) syndrome. This clinical observation led to the search for the molecular player implicated in RIDDLE syndrome. In 2009, several independent studies demonstrated that inactivation of RNF168 leads to defective 53BP1 repair foci formation, which also links to the RIDDLE syndrome pathology [17–19]. Four cases of RIDDLE syndrome have been reported to have frameshift mutations within RNF168 resulting in loss-of-function truncations (illustrated in Fig.1): c.397dupG/c.1323_1326delACCA, c.391C>T homozygous which result in A133G/N441R frameshift mutations; R131X [18, 21]; and two cases of homozygous c295delG which leads to E99K frameshift mutation [22]. These observations identified RNF168 as the key player in RIDDLE syndrome pathology.
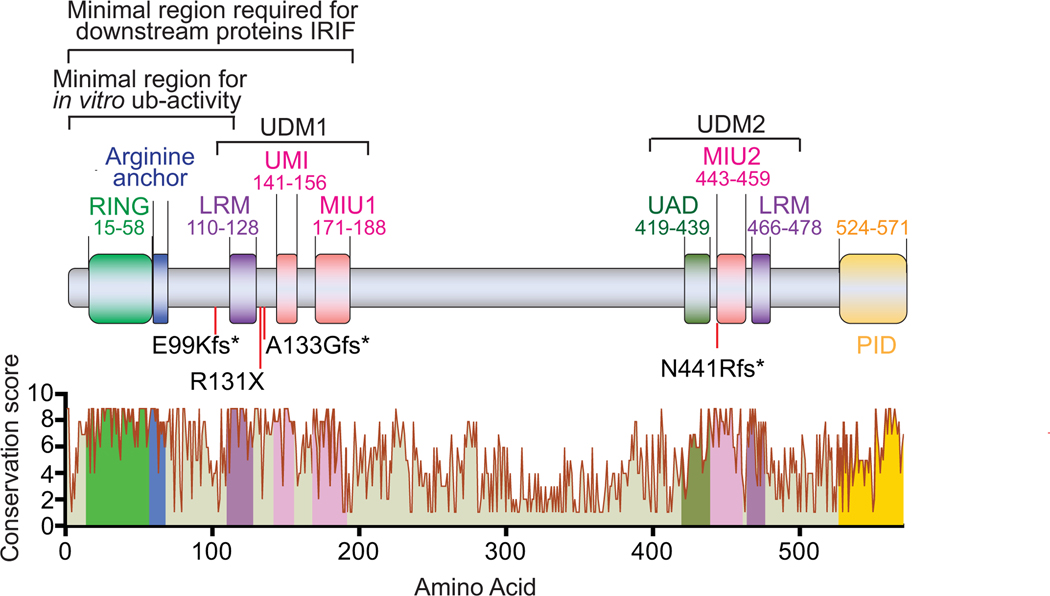
Figure 1. Human RNF168 (RIDDLIN), an evolutionarily conserved ubiquitin E3 ligase.
RNF168 is composed of a catalytic RING domain and two ubiquitin-dependent DSB recruitment modules (UDM). UDM1 is composed by LRM, UMI and MIU1 and UDM2 is composed by UAD, MIU2 and LRM. RNF168 also contains an arginine anchor for substrate recognition. Corresponding conservation score was analyzed using the ConSurf Server: https://consurf.tau.ac.il. Conservation score is calculated using an empirical Bayesian methodology by comparing 107 vertebrates that share 35–95% homology. The scores are normalized and grouped into nine conservation grades [23]. Mutations found in patients with RIDDLE syndrome are marked on the schematic illustration of RNF168 as E99Kfs*, R131X, A133Gfs* and N441Rfs*. LRM (LR motif); UMI (UIM: ubiquitin interacting motif- and MIU: motif interacting with ubiquitin-related ubiquitin-binding motif) and MIU (Motif interacting with ubiquitin); UAD (ubiquitin associated domain); PID (PALB2 interacting domain).
2. Structural architecture and functional domains of RNF168
The structural and functional domains of RNF168 have been well defined and extensively characterized. RNF168 contains a RING domain at the N-terminus, a ubiquitin-dependent DSB recruitment module (UDM)1 in the middle and a UDM2 in the C-terminus. Human RNF168 protein sequence analysis across 107 vertebrates that share 35%−95% homology showed that the functional domains of RNF168 are highly evolutionarily conserved (Fig. 1) [23].
The catalytic RING domain at the N-terminus (a.a. 15–58) confers ubiquitin E3 ligase activity as well as substrate specificity for downstream functions [24, 25]. The C-terminus of the RING domain includes an arginine-rich region, which serves as an anchor for the nucleosome acidic patch, facilitating H2A ubiquitination. Mutations of the arginine anchor in RNF168 or the acidic patch in the nucleosome largely abolish the H2A(X) K13/K15 ubiquitination [26, 27]. The crystal structure of the RNF168 RING finger has been solved [24, 28]. The catalytic cysteine residues and arginine anchors are highly conserved, which is demonstrated by the analysis of the RING domain structure (PDB: 4gb0) by comparing 107 vertebrates that share 35–95% homology (Fig. 2A).
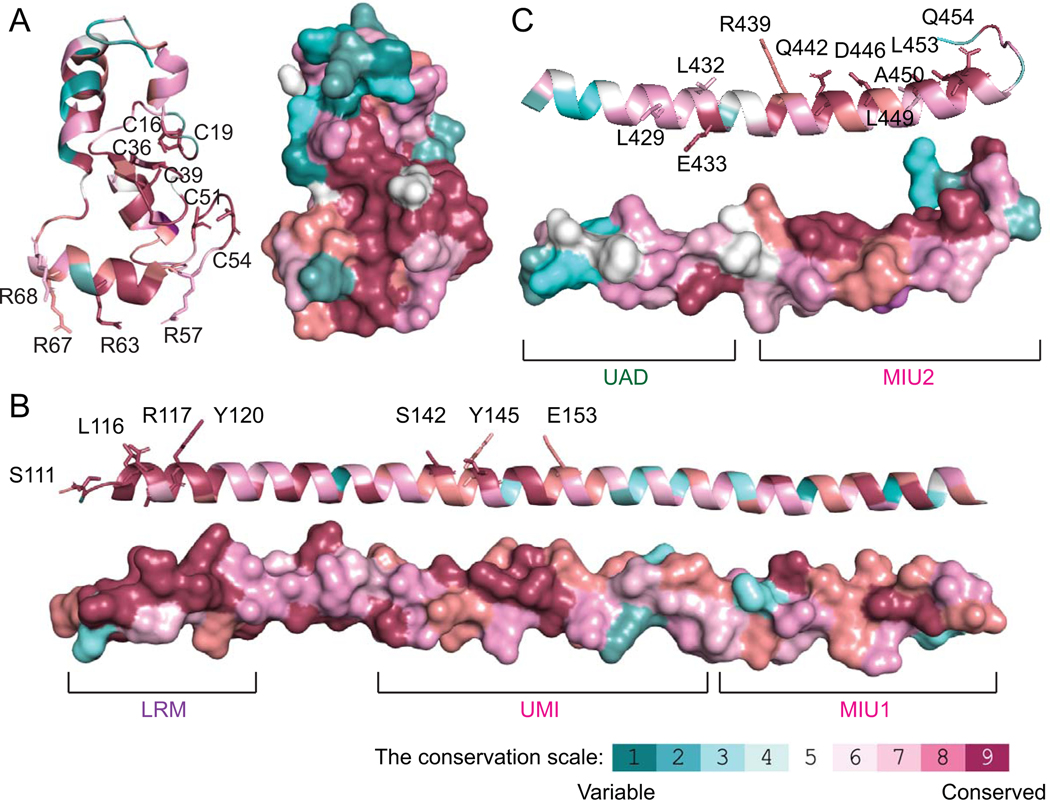
Figure 2. Conservation of the RNF168 functional domains.
Evolutionary conservation scores were analyzed by the ConSurf Server and pseudo-colored to indicate amino acid conservation. A) Ribbon (left) and space-filling (right) models of the RNF168 RING domain crystal structure (PDB ID:4GB0). B) Ribbon (Top) and space-filling (bottom) of the RNF168 UBD1 (PDB ID:5XIS) crystal structure. C) Ribbon (Top) and space-filling (bottom) of the RNF168 UDM2 (PDB ID: 5XIU) crystal structure. Labeled sticks indicate the characterized functional residues.
The UDM1 is comprised of three motives: LRM (LR motif) 1, UMI (UIM: ubiquitin interacting motif- and MIU: motif interacting with ubiquitin-related ubiquitin-binding motif) domain, and MIU (motif interacting with ubiquitin) 1. The LRM1 (a.a. 110–128) is essential for directing the UMI and MIU1 to ubiquitinated ring finger protein 8 (RNF8) [29]. The UMI (a.a. 141–156) preferentially binds to K63-linked ubiquitin chains. Inactivation of K63 binding partially impairs the subcellular localization of RNF168 upon DNA damage [30]. The essential residues and ubiquitin binding amino acids are highly conserved within this region (Fig. 2B). The MIU1 (a.a.171–188) juxtaposes with the UMI domain. Mutation of the MIU1 motif drastically reduces RNF168 binding to K48 ubiquitin chains [30]. However, a recent study showed that the MIU1 domain alone exhibits a similar binding affinity to mono-ubiquitin, K48, and K63 ubiquitin chain [31]. Interestingly, MIU1 deletion exhibits modest impairment of RNF168 ionizing radiation-induced foci (IRIF) that are generated by the sequential assembly of DNA repair proteins at DSBs but does not inhibit the recruitment of downstream repair proteins to damaged chromatin [18]. The UMI/MIU1 is responsible for the localization of RNF168 to promyelocytic leukemia (PML) nuclear bodies, which promotes ubiquitination and sumoylation of PML bodies. Together, the UDM1 preferentially binds to K63-linked ubiquitin chain. It functionally acts downstream of RNF8 by interacting with RNF8 substrates, which triggers the initial localization of RNF168 at DNA breaks [29].
The UDM2 contains a UAD (ubiquitin-associated domain), an MIU2 and a LR motif [29, 31]. The UAD (a.a. 419–439) has been recently identified as an essential entity for the distal K63 chain recognition [31]. The biological role of the UAD is yet to be characterized. The highly evolutionarily conserved MIU2 (a.a. 443–459) domain is located at the C-terminus of RNF168 (Fig. 2C). Deletion of MIU2 motif drastically reduces the RNF168 IRIF formation [17, 18]. Recent studies showed that MIU2 has micromolar range binding affinity to H2A(X) K15 ubiquitination [32], suggesting that RNF168 generates a positive feedback ubiquitination signal to and for its localization at damaged chromatin, thus establishing a strong local ubiquitin signal and facilitating the robust recruitment of 53BP1 to damaged chromatin [29].
The LRM2 (a.a. 466–478) shares minimal homology with LRM1. These two LRMs carry out different functions in regulating RNF168 at damaged chromatin. LRM1 targets the upstream protein(s) that are ubiquitinated by RNF8, whereas LRM2 serves as a nucleosome binding motif to enhance the MIU2 interaction with the RNF168-mediated H2A(X) K15 ubiquitination. The LRM2 promotes the MIU2-mediated RNF168-self amplification of accrual at damaged chromatin via interaction with the H2A(X) K15 ubiquitination [29]. In line with these observations, overexpression of RNF168 lacking UDM2 restores downstream DDR protein foci formation in RNF168-depleted cells [25, 33]. Taken together, the UDM2 serves as a rate-limiting self-amplification machinery to trigger the RNF168-mediated ubiquitin signaling pathway in the proximity of DNA breaks.
Moreover, although the RNF168 C-terminus is not a putative structural domain, it interacts with Partner and localizer of BRCA2 (PALB2) and promotes RAD51 accrual at DSBs. The interaction confers the potential regulatory role of RNF168 in HR repair [34]. The RNF168-mediated PALB2 loading on damaged DNA supports Breast cancer susceptibility gene 1 (BRCA1)-independent HR repair. This “backup” HR pathway in BRCA1 haploinsufficiency requires chromatin ubiquitination [35]. Thus, the RNF168 domains neatly collaborate at DSBs to precisely execute its functions in orchestrating the DDR pathway.
3.1. RNF168 and the DDR pathway
RNF168 has been identified as a key player in manifesting the ubiquitin signaling in the DDR pathway. The E3 ligase activity of RNF168 promotes different ubiquitin signaling entities: mono-ubiquitination, K48-linked, K63-linked and K27-linked ubiquitin chains [12, 36] to regulate DNA damage signaling.
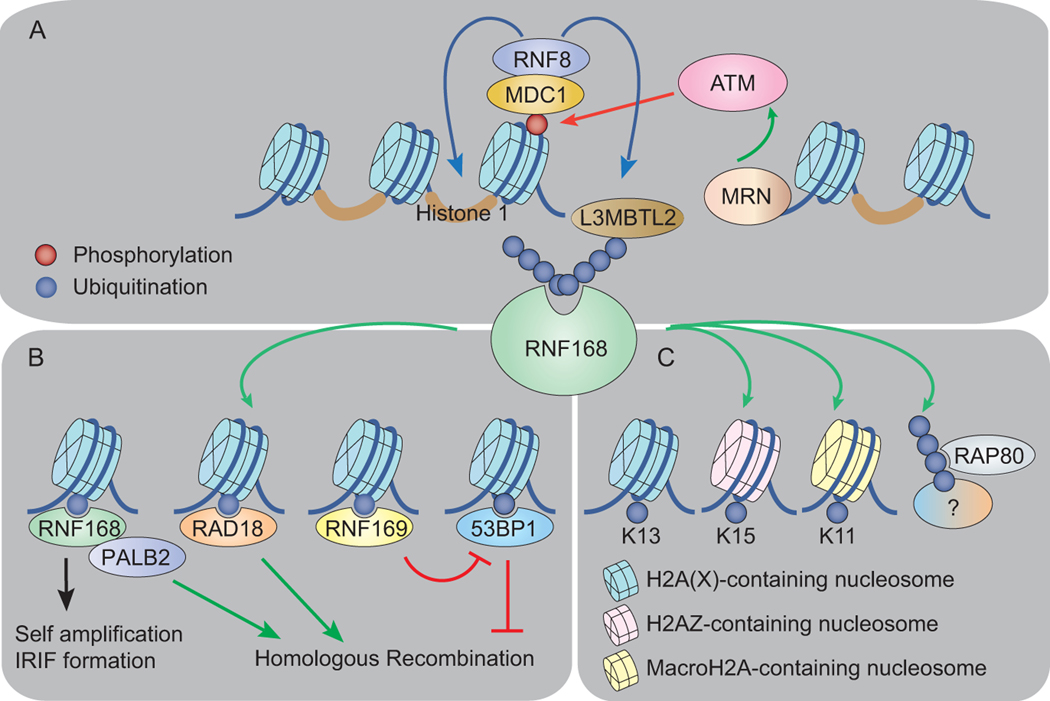
DNA DSBs are sensed by MRE11-RAD50-NBS1 (MRN) complex, which activates ataxia telangiectasia mutate (ATM) kinase [37, 38]. The initial sensing, recognition and kinase activation mechanism is evolutionarily conserved [39]. ATM kinase phosphorylates H2AX at serine 139 (γH2AX), which marks the sites of DSBs. γH2AX provides a docking platform for mediator of DNA damage and checkpoint 1 (MDC1) binding. Subsequently, RNF8 is recruited to DSBs via protein-protein interaction with MDC1 [4, 40]. Initial DSB recruitment of RNF168 depends on its physical interaction with the RNF8 substrates histone 1.2 and Lethalmalignant brain tumor-like 2 (L3MBTL2) [41, 42] (Fig. 3A).
Figure 3. RNF168 as a key DNA repair molecular switch on damaged chromatin.
A) A simplified overview of the genetic regulation of RNF168 accrual at damaged chromatin. B) The RNF168-mediated H2A(X) K15 ubiquitination and the characterized potential readers that are involved in DNA repair. C) The unsolved riddle of RNF168. Schematic representation of the uncharacterized RNF168 substrates H2AZ and macroH2A and the unidentified RNF168 substrate that regulates RAP80 and BRCA1 IRIF.
RNF168 is considered as the rate limiting determinant for the recruitment of key DDR proteins, including 53BP1, BRCA1 and the E3 ubiquitin protein ligase RAD18, to damaged chromatin. RNF168 level is tightly controlled by TRIP12 and UBR5 to prevent excessive amplification of the ubiquitin signal [43].
The initial recruitment of RNF168 to damaged chromatin triggers site-specific H2A(X) mono-ubiquitination as well as K63-linked ubiquitin conjugates at K13 (H2A(X)K13ub) and K15 (H2A(X)K15ub), a prerequisite for DSB repair [19, 44, 45]. While the role of H2AK13ub is largely unclear, the H2A(X)K15ub serves as a recruitment scaffold for the localization of ubiquitin binding DNA repair proteins at damaged chromatin, which include 53BP1 via its ubiquitin-dependent recruitment (UDR) motif [46]. The DSB recruitment of 53BP1 promotes NHEJ by inhibiting DNA end resection, the first step of HR [6, 47]. RNF168 catalyzes a K63-linked ubiquitin chain on 53BP1 at K1268 to promote its initial DSB recruitment [48] and on TOP2 regulating its chromatin binding and decatenation activity [49]. In addition, RNF168 was also found to catalyze K48-linked ubiquitin chain specifically on forkhead box M1 (FOXM1) in breast cancer cells, leading to FOXM1 proteasomal degradation upon epirubicin treatment [50]. In cooperation with RNF8, RNF168 also promotes the K48-linked ubiquitination and subsequent proteasomal degradation of KDM4A/JMJD2A, a dimethylated H4K20 reader. The removal of KDM4A/JMJD2A H4K20me2 occupancy facilitates 53BP1 DSB recruitment [51]. In addition to canonical K63- and K48-linked ubiquitin chains, RNF168 also catalyzes the non-canonical K27-linked ubiquitin chain at chromatin. Proteomic analysis shows that H2A(X) K13/K15 are the target substrates of K27 ubiquitin conjugates. K27 ubiquitination is indispensable for proper activation of the DDR and repair protein assembly at DSBs [36].
Other DNA repair proteins also show comparably high affinity to H2A(X)K15ub, which include RAD18 via its ubiquitin zinc finger (UBZ) domain; RNF169 via its UDM2 domain [32]; and BRCA1 associated RING domain 1 (BARD1) via its tandem BRCA1 C-terminal (BRCT) domain [52]. The H2A(X)K15ub also interacts with the RNF168 ubiquitin binding domain 2 (UBD2), positively amplifying RNF168 IRIF at DSBs [29, 32]. Recently, RNF168 was found to ubiquitinate H2AZ and macroH2A histone variants in vitro and in vivo [25]. However, structural and biological implications of their ubiquitination remain to be examined.
Overall, current data strongly suggest that RNF168 is the molecular switch for the DDR (Fig. 3B). Emerging evidence further supports the notion that the DNA repair proteins directly interact with the H2A(X)K15ub mark. The choice of DSB repair pathway is likely executed by co-operating with a secondary histone mark orchestrating the assembly of distinct repair protein complexes at DSB sites throughout the cell cycle.
In addition to site-specific H2A ubiquitination, the RNF168-catalyzed K63-linked ubiquitin chain is responsible for recruiting the BRCA1-A complex [53]. The BRCA1-A complex is composed of seven proteins, including BRCA1-BARD1, RAP80, ABRA1, MERIT40, BRCC36, and BRE. RAP80 acts most upstream of this complex, promoting recruitment of BRCA1-A to DSB sites. It contains a UIM which interacts with the K63-linked ubiquitin chains potentially generated by RNF168. BRCC36 is a deubiquitinase (DUB) that cleaves K63-linked ubiquitin chains, while ABRA1, MERIT40 and BRE play a role in BRCA1-A complex stabilization (see reviews [54, 55]). The BRCA1-A complex prevents hyperactive HR by suppressing DNA end resection [56, 57]. One model suggests that the BRCA1-A complex sequestrates the BRCA1-HR function from DNA end processing [58]. Nevertheless, BRCA1/BARD1 catalytic function is not well-characterized within this complex.
In general, RNF168-mediated H2A(X) K13/K15 ubiquitination promotes NHEJ and suppresses HR function [6, 59, 60], potentially through recruiting 53BP1 to DSBs to suppress DNA end resection. It also can sequester BRCA1 HR function or HR function of BRCA1 via RAP80 binding to an unidentified K63 conjugated substrate. RNF168 was also shown to be involved in single-strand annealing defects caused by BRCA1-depletion [60].
The mechanism by which RNF168 regulates NHEJ is well-studied. Paradoxically, RNF168 is also involved in in promoting homology-directed repair, which is demonstrated by its indispensable role of recruiting PALB2 and RAD18 to DNA damage [34, 61]. Recent studies show that RNF168 supports HR independent of BRCA1 via PALB2 and RAD51 loading at damaged chromatin. Interestingly, a subset of BRCA1-mutant cells display reduction in RNF168 protein levels and genetic ablation of RNF168 reveals BRCA1 haploinsufficiency [35, 62]. Together, these data support the role of RNF168-mediated ubiquitination as a backup mechanism for HR in the absence of BRCA1. Collectively, RNF168 is a multifaceted regulator of the intricately orchestrated DDR pathway at damaged chromatin.
3.2. RNF168 and the replication fork
RNF168 is localized at the replication fork and is required for efficient DNA replication and fork progression via H2A ubiquitination [63]. RNF168 depletion leads to repetitive sequence replication defects and chromosomal abnormalities. Interestingly, ectopic RNF168 expression recruits RAD18-SLF1 via γH2AX K15 ubiquitination and subsequently leads to break-induced replication in the absence of homologous recombination at stalled replication forks [64]. RNF168-driven break-induced replication may contribute to the mutational signatures in BRCA1-mutated genomes. Together, these observations suggest that tight regulation of RNF168 expression and localization are essential for replication fork stability in physiological and pathological conditions.
3.3. Viral interaction with RNF168
The DDR pathway has been shown to be involved in the cellular response to viral infection [65]. In particular, the ubiquitin pathway is one of the major targets for viral proteins to block immune response and to enhance viral fitness by promoting virus replication, relief of transcriptional repression, and activation of latent viral genomes [66]. For instance, herpes simplex virus E3 ubiquitin ligase ICP0 targets and degrades RNF8 and RNF168 to suppress H2A(X) ubiquitination and inhibit the DDR pathway [67].
Interestingly, human papillomaviruses (HPV) hijack the DDR pathway to promote the viral replication cycle. A subset of human HPV-positive tumors displays increased RNF168 protein levels [43, 68]. The HPV early protein E7 interacts with RNF168 at the region between MIU1 and MIU2. The interaction does not affect the enzymatic activity for RNF168. However, the interaction of HPV E7 sequestrates RNF168 and interferes with DNA repair pathway choice by impairing ubiquitin signaling and 53BP1 damage-induced foci formation. Consequently, HPV E7 promotes genome instability and drives the progression of cervical and head and neck cancers [68]. Overall, oncogenic viruses could potentially target RNF168 and subvert the DDR pathway to expand the lifespan of host cells. Thus, inhibition of the interaction between RNF168 and virus could be used as a therapeutic target to suppress virus replication and maintain genome stability,
4. RNF168 substrate specificity
The upstream genetic pathway and regulatory mechanism for RNF168 has been well characterized over the last decade. However, the mechanistic underpinnings of RNF168 substrate recognition and interaction are still an active area of research [26, 27, 31].
The RNF168 RING domain is functionally non-transferrable. Domain swapping shows that replacement of the RNF168 RING domain with the RNF8 RING domain does not restore IRIF formation of FK2 (ubiquitin) [69] or the DDR proteins 53BP1 and BRCA1 in RNF168-depleted cells or RIDDLE cells [24, 25]. Similarly, replacing the RNF8 RING domain with the RNF168 RING domain does not restore FK2 foci or DDR repair protein foci in RNF8-depleted or RNF168-depleted cells, despite its ability to localize at damaged chromatin [25]. These observations suggest that the RNF168 RING domain confers E3 ligase activity and determines substrate specificity.
RNF168 RING-arginine anchor fragment (a.a. 1–113) is sufficient for H2A(X) ubiquitination in vitro (Fig. 1) [26]. However, RNF168 RING-arginine anchor overexpression or tethering this fragment at DNA breaks by substitution of the RNF8 RING domain with RNF168 RING-arginine anchor does not rescue the DDR foci in RNF168-depleted cells regardless of its ability to form IRIF [25].
Moreover, the RNF168 fragment consisting of RING-UMI-MIU1 (a.a. 1–189) shows a noticeably higher activity to H2A(X) in vitro [26]. Reconstitution of an RNF168 fragment harboring the RING-UMI-MIU1 (a.a. 1–190 or a.a. 1–220) restores the recruitment of DDR proteins to IRIF in RNF168 knockout (KO) cells despite its inability to form IRIF (Fig. 1) [25, 33]. Together, these data further support the possibility that other sequence determinants within a.a. 113–190 beside the RING domain mediate RNF168-substrate recognition [14].
RNF168 ubiquitinates H2A, H2AX, H2AZ, and macroH2A1/2 at their N-terminus (H2A(X): K13 and K15, H2AX K15, macroH2A1/2 K11). The target recognition is restricted within three amino acids at the N-terminus of the alpha1-extension helix [25]. Ectopic RNF168 expression does not show non-specific ubiquitination to any other lysine residues on the nucleosome, which contains more than a hundred lysine residues. Overall, the targeting specificity is extremely constrained suggesting that the RNF168-mediated DDR pathway is highly regulated.
The detailed molecular mechanism by which RNF168 achieves its target specificity toward H2A variants N-terminus in the context of the nucleosome has been progressively unveiled. It utilizes the arginine stretch at the C-terminus of the RING domain to anchor the nucleosome at its acidic patch. This molecular interaction is established via the electrostatic potential difference. The arginine anchor (R57, R63, and R67) of RNF168 forms hydrogen bonds with the nucleosome acidic patch (E60, E63, D89, and E91) [27].
Additionally, the evolutionarily and structurally conserved H2A N-terminal alpha1-extension helix is required for RNF168-mediated site-specific ubiquitination without perturbing the RNF2-mediated ubiquitination at the H2A C-terminus. The nucleosome shows an opposing discrepancy in electrostatic potential between the H2A N-terminal and C-terminal tails, which potentially contributes to the precise orientation of RNF168 on the nucleosome for target residues specificity [25]. The alpha1-extension helix may partially interact with nucleosomal DNA. However, RNF168 can catalyze H2A ubiquitination in the context of H2A/H2B dimer, which rules out the possibility that the abrogation of RNF168-mediated H2A ubiquitination after alpha1-extension helix mutation is due to nucleosome destabilization
Interestingly, the region between the RING domain and MIU1 is evolutionarily acidic [25, 41]. Comprehensive mapping demonstrated that the UMI E143/E144 is potentially required for targeting the positively charged H2A N-terminal nucleosome surface [25]. In conjunction with the RNF168 arginine anchor and nucleosome acidic patch binding, the potential interaction between the RNF168 E143/E144 and H2A alpha1-extension helix creates a unidirectional orientation for RNF168 to execute the site-specific ubiquitination. It is unlikely the UMI E143/E144 residues affect the ubiquitin binding affinity as it is not located on the ubiquitin-binding interface of the UMI domain [30]. Together, current evidence suggests RNF168 requires more than one substrate-recognition entity to accomplish its molecular target selectivity. Structural studies will provide further insight into the molecular mechanism.
Overall, understanding the molecular regulation of RNF168 substrate recognition will provide important insights into identifying novel RNF168 substrates and delineating the downstream regulatory mechanisms of DDR-ubiquitin signaling.
5. Counteracting mechanisms for RNF168-mediated DDR pathway
Deubiquitinases (DUBs) turn off the RNF168 E3 ligase-mediated signals to strike a balance during DNA damage signaling. Multiple H2A DUBs, including ubiquitin-specific proteases USP3, USP16, USP26, USP37, USP44, and USP51 have been identified as negative regulators for RNF168-mediated H2A(X) K15 ubiquitination.
USP3 depletion in both human cells and mouse models show global elevation of H2A ubiquitination, 53BP1 foci, checkpoint dysregulation, genome instability, and spontaneous tumor development [70, 71]. Conversely, overexpression of USP3 reduces H2A(X) ubiquitination, as well as RNF168, 53BP1, RAP80, and BRCA1 IRIF formation [17, 72, 73]. Overall, despite the non-specificity towards H2A ubiquitination [74], current data support the notion that USP3, at least in part, counteracts the RNF168-mediated pathway in regulating the DDR.
USP16 specifically deubiquitinates H2A on both DNA damage-induced N-terminal K13/K15 and C-terminal K118/K119 in vivo and in vitro. Depletion of USP16 increases FK2 foci. Although USP16 overexpression is largely localized in the cytoplasm, it impairs FK2 and 53BP1 IRIF without affecting the RNF8 and RNF168 IRIF upon DNA damage [75, 76].
USP44 was found to counteract RNF8/RNF168-dependent histone ubiquitination at DSBs and is localized at DSBs in an RNF168-dependent manner. Ectopic USP44 overexpression suppresses 53BP1 recruitment to damaged chromatin [73].
Overexpression of either USP26 or USP37 inhibits 53BP1 IRIF formation [73, 77]. They also suppress the excessive spreading of RAP80-BRCA1 from DSBs by removing RNF168-induced distal ubiquitin conjugates from DSBs [77]. USP26 and USP37 limit the ubiquitin-dependent sequestration of BRCA1 and facilitate BRCA1-PALB2-BRCA2-RAD51 association at damaged chromatin to promote HR repair [77]. However, it is unclear whether they target H2A(X)K15ub directly.
USP51 is a site-specific deubiquitinase for H2A K13/K15 in vitro [78]. Overexpression of USP51 suppresses H2A ubiquitination as well as 53BP1 and BRCA1, but not RNF168 IRIF formation. USP51 depletion leads to ionizing radiation hypersensitivity and dysregulated DNA repair [78].
Several studies report potential DUBs, including USP10, USP11, USP29, OTUB1, and BRCC36, negatively regulate the RNF168-associated ubiquitin signaling [49, 73, 79, 80]. Nevertheless, detailed mechanisms, substrate specificity, and genetic regulation have not been established. Further investigation is needed to understand their roles in the RNF168-mediated ubiquitin signaling DNA repair pathway.
The collaboration of the DUBs plays an important role in preventing excessive spreading of ubiquitin signals at damaged chromatin that are mediated by RNF8 and RNF168. Some DUBs enable modulation of specific downstream complexes of RNF168. Overall, this mechanism strikes a balance to properly repair the DNA break in a timely manner.
6. The RNF168-dependent crosstalk between phosphorylation and ubiquitination
The interplay between ubiquitination and phosphorylation has been an exciting area of research. It has been suggested that ubiquitin can be phosphorylated at almost every serine, threonine, and tyrosine residue [81, 82]. However, the functional consequences and the regulation of ubiquitin phosphorylation remain largely unexplored. A recent study showed that the ubiquitin molecule conjugated on H2A(X)K15 can be phosphorylated at the threonine (T) 12 residue (pUbT12) in DNA damage in an RNF168-dependent manner [83]. pUbT12 localizes at DNA damage and inhibits 53BP1 accessibility to H2A K15ub but not HR promoting factors such as RNF169 and BARD1/BRCA1. This phosphorylation-ubiquitination crosstalk enables an additional layer of regulation to direct the cells to HR repair.
7. Unsolved RIDDLES
RNF168 is a central regulator at the crossroads of the DDR pathway. Present evidence strongly supports that this ubiquitin signaling step is essential for bringing many of the major DNA repair factors, including 53BP1, BRCA1, and RAD18, to the DNA breaks.
Remarkable progress has been made over the last decade in understanding the mechanistic regulation on how RNF168 recruits 53BP1 to damaged chromatin via its substrate H2A(X). 53BP1 UDR domain has a high specificity for the ubiquitinated K15 of H2A(X) and its surrounding sequence. It is also largely selective to the sequence flanking the target ubiquitinated lysine [46]. Interestingly, the H2A(X) K15 ubiquitination mark shows a high affinity to the RNF168 MIU2 motif, RAD18 UBZ domain, RNF169 MIU2 domain, and BARD1 tandem BRCT domain. These proteins function differentially in the context of the DDR, which prompted a peculiar paradox of how exactly they come into place to refine the DNA repair pathway choice throughout the cell cycle. One possibility is that there may be additional histone marks or regulatory proteins involved in stabilizing and destabilizing their interactions with damaged chromatin in cells. Moreover, the biological function of H2A(X)K13ub is uncharacterized.
RNF168 utilizes the MIU2-H2A(X) K15 ubiquitination interaction to self-amplify at DNA breaks to open additional ubiquitin docking platforms for downstream repair proteins at damaged chromatin [29, 32]. However, it is currently unclear why RAD18 and RNF168 compete for the same docking platform with 53BP1 in vivo.
Additionally, RAP80 is the upstream regulator of BRCA1 IRIF formation. Although BARD1 promotes BRCA1 DSB recruitment, the genetic and mechanistic regulation of the RNF168-RAP80-BRCA1 axis is not fully delineated. There is a possibility that an unidentified RNF168 substrate mediates RAP80 DSB recruitment.
Based on sequence analysis, it is unlikely that the 53BP1 UDR binds RNF168-mediated ubiquitinated H2AZ and macroH2A1/2, which raises the question of what are the functions of specific histone marks in the context of DNA repair? One of the current predicting models is that H2A variants occupy and concentrate at different genomic loci and their ubiquitination provides a specific docking platform for different repair proteins to repair the damaged DNA in different chromatin states. Overall, given the currently established roles of H2AZ and macroH2A in maintaining genome stability and the DDR regulation [84–88], dissecting the biological implication of H2AZ K15 and macroH2A1/2 K11 ubiquitination is important for understanding the regulation of DNA damage signaling in the context of damaged chromatin modification.
Interestingly, RNF168 MIU2 has high affinity for its target substrate. It is not completely elucidated how the MIU2-H2AX K15 ubiquitination interaction self-amplifies RNF168 at damaged chromatin. One possibility is that the binding facilitates an allosteric change of the RNF168 RING domain and repositions the RNF168 N-terminus to the next nucleosome, which allows for a rapid spread of ubiquitin signal at the DNA break by retaining RNF168 in the proximity of the DNA breaks.
The recent discovery of the RNF168 substrate recognition entities [25, 26, 89], will help identify additional RNF168 substrates in the context of the DDR pathway. Specifically, proteomic or bioinformatic approaches on identification of the nucleosome acidic patch-RNF168 arginine anchor and H2A alpha1-extension helix-RNF168 UIM may shed light on revealing the RNF168-downstream network.
Studies show that the chromatin-based DDR pathway is not evolutionarily conserved. The functional H2AX-MDC1-RNF8-RNF168 axis is not present in yeast or prokaryotes. In particular, mdb1 (the fission yeast homolog of human MDC1) and Dma (the budding yeast homolog of RNF8) do not recapitulate the human homolog’s phenotype [90, 91]. Budding and fission yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe) have an hH2AX homolog, yeast HTA (yHTA), but lack an RNF168 homolog. Interestingly, 53BP1 homologs in yeast: rad9 in Saccharomyces cerevisiae and crb2 in Schizosaccharomyces pombe, do not seem to have a UDR domain, suggesting that rad9 and crb2 are regulated by a different mechanism at damaged chromatin independent of the ubiquitin signaling. There are two distinctive evolutionarily differences between H2AX and yHTA at their histone tails. First, H2AX C-terminus harbors the -SQEY motif which is required for MDC1 binding, whereas the yHTA C-terminal -SQEL has been shown to be defective in binding MDC1 [92]. Although it is unclear if the mdb1-yHTA interaction requires the leucine residue at the very C-terminus, the recruitment of rad9 or crb2 to DNA breaks is unlikely to be mdb1-dependent. Second, RNF168 does not ubiquitinate yHTA despite the presence of lysine residues at the N-terminus. It is due to the RNF168-mediated H2A(X) ubiquitination that it is restricted by the distance between the target lysine residue(s) and the alpha1-extension helix. The most proximal yHTA N-terminal lysine from the alpha1-extension helix is four amino acids apart. These observations suggest that the H2A histone tails might have co-evolved with MDC1, RNF168 and 53BP1 to achieve an additional layer of ubiquitin signaling regulation to cope with the increased complexity of DNA repair in eukaryotes. Further analysis will provide more insights into the evolution of the chromatin-based DDR pathway.
8. Concluding remarks
Defective DNA repair-manifested genomic instability is a common driving force of multiple types of cancers. The DDR pathway provides attractive targets for therapeutic interventions based on the synthetic lethality paradigm. One of the best-exploited examples is the use of PARP inhibitors (PARPi) to inactivate base excision repair for treating homologous recombination repair defective cancers, particularly those with BRAC1 or BRCA2 mutations. Although there are only four RIDDLE syndrome cases reported with RNF168 mutations, dysregulation of RNF168 is implicated in tumorigenesis. More specifically, several lines of evidence show that RNF168 is intimately linked to cancer subtypes [33, 62, 93]. Cells with elevated RNF168 are more resistant to chemotherapy and radiotherapy. The aberrant RNF168 signaling pathway may lead to genotoxic stress adaptation [93]. Thus, RNF168 protein level in cancer patients can be used to predict therapeutic response. Given that RNF168 plays an essential role in BRCA1 haploinsufficient cells, targeting RNF168 or chromatin ubiquitination could be a therapeutic strategy to kill cancer cells, particularly in BRCA1-deficient cells that are resistant to PARPi [35]. Identification of RNF168-substrates regulatory elements, such as the nucleosome acidic patch and the H2A alpha1-extension helix, provides additional targets for therapeutic development [25, 26, 89].
The DDR pathway contains druggable targets for cancer therapy. Ubiquitination is one of the most important epigenetic switches in cellular functions, including the DDR pathway. Hence, it is imperative to understand the regulation of this essential biological process. With multiple regulatory domains, RNF168 is an attractive candidate for therapeutic intervention that remains to be explored.
Acknowledgements
J.L. is supported by grants from NIH (NCI: K22CA204354, R01CA244261, NIGMS: R35GM137798, and P20GM121293; J.L. as project leader), Arkansas Breast Cancer Research Program (AWD00053730 and AWD00054499) and American Cancer Society (RSG-20–131-01-DMC). G.G is supported by NIGMS COBRE (P20GM121316; G.G. as project leader and NCI K22CA188181). We thank Halle Mallard for proofreading the manuscript. We apologize to all the studies we did not reference due to space constraints.
Abbreviations
- RIDDLE
Radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties
- RNF168
RING finger protein 168
- E1
ubiquitin activating enzyme
- E2
ubiquitin conjugating enzyme
- E3
ubiquitin ligase
- IRIF
Ionizing radiation-induced foci
- DDR
DNA damage response
- NHEJ
Non-homologous end joining
- HR
Homologous recombination
- DSB
Double strand break
- 53BP1
tumor suppressor p53 binding protein 1
- BRCA1
Breast Cancer susceptibility gene 1
- UDM
ubiquitin-dependent DSB recruitment module
- UDR
Ubiquitin-dependent recruitment
- DUB
Deubiquitinase
- UIM
ubiquitin interacting motif
- MIU
motif interacting with ubiquitin
- UMI
UIM and MIU related ubiquitin-binding motif
- UAD
ubiquitin associated domain
Footnotes
Conflicts of Interest
The authors declare no conflict of interest
References
- 1.Jackson SP & Bartek J. (2009) The DNA-damage response in human biology and disease, Nature. 461, 1071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccia A & Elledge SJ (2010) The DNA damage response: making it safe to play with knives, Mol Cell. 40, 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polo SE & Jackson SP (2011) Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications, Genes Dev. 25, 409–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scully R, Panday A, Elango R & Willis NA (2019) DNA double-strand break repair-pathway choice in somatic mammalian cells, Nat Rev Mol Cell Biol. 20, 698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symington LS & Gautier J. (2011) Double-strand break end resection and repair pathway choice, Annu Rev Genet. 45, 247–71. [DOI] [PubMed] [Google Scholar]
- 6.Chapman JR, Taylor MR & Boulton SJ (2012) Playing the end game: DNA double-strand break repair pathway choice, Mol Cell. 47, 497–510. [DOI] [PubMed] [Google Scholar]
- 7.Kanaar R, Wyman C & Rothstein R. (2008) Quality control of DNA break metabolism: in the ‘end’, it’s a good thing, EMBO J. 27, 581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.House NC, Koch MR & Freudenreich CH (2014) Chromatin modifications and DNA repair: beyond double-strand breaks, Front Genet. 5, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dantuma NP & van Attikum H. (2016) Spatiotemporal regulation of posttranslational modifications in the DNA damage response, EMBO J. 35, 6–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukas J, Lukas C & Bartek J. (2011) More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance, Nat Cell Biol. 13, 1161–9. [DOI] [PubMed] [Google Scholar]
- 11.van Attikum H & Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response, Trends Cell Biol. 19, 207–17. [DOI] [PubMed] [Google Scholar]
- 12.Uckelmann M & Sixma TK (2017) Histone ubiquitination in the DNA damage response, DNA Repair (Amst). 56, 92–101. [DOI] [PubMed] [Google Scholar]
- 13.Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK & Durocher D. (2010) The ubiquitous role of ubiquitin in the DNA damage response, DNA Repair (Amst). 9, 1229–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swatek KN & Komander D. (2016) Ubiquitin modifications, Cell Res. 26, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akutsu M, Dikic I & Bremm A. (2016) Ubiquitin chain diversity at a glance, J Cell Sci. 129, 875–80. [DOI] [PubMed] [Google Scholar]
- 16.French ME, Koehler CF & Hunter T. (2021) Emerging functions of branched ubiquitin chains, Cell Discov. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J & Lukas C. (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins, Cell. 136, 435–46. [DOI] [PubMed] [Google Scholar]
- 18.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM & Durocher D. (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage, Cell. 136, 420–34. [DOI] [PubMed] [Google Scholar]
- 19.Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G & Penengo L. (2009) RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX, BMC Mol Biol. 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, Drayson MT, West SC, Elledge SJ & Taylor AM (2007) RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling, Proc Natl Acad Sci U S A. 104, 16910–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devgan SS, Sanal O, Doil C, Nakamura K, Nahas SA, Pettijohn K, Bartek J, Lukas C, Lukas J & Gatti RA (2011) Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia, Cell Death Differ. 18, 1500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietrucha B, Heropolitanska-Pliszka E, Geffers R, Enssen J, Wieland B, Bogdanova NV & Dork T. (2017) Clinical and Biological Manifestation of RNF168 Deficiency in Two Polish Siblings, Front Immunol. 8, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T & Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules, Nucleic Acids Res. 44, W344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Chen J, Wu M, Wu H, Arokiaraj AW, Wang C, Zhang W, Tao Y, Huen MS & Zang J. (2013) Structural basis for role of ring finger protein RNF168 RING domain, Cell Cycle. 12, 312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelliher JL, West KL, Gong Q & Leung JWC (2020) Histone H2A variants alpha1-extension helix directs RNF168-mediated ubiquitination, Nat Commun. 11, 2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ & Sixma TK (2014) The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A, Nat Commun. 5, 3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn V, Uckelmann M, Zhang H, Eerland J, Aarsman I, le Paige UB, Davidovich C, Sixma TK & van Ingen H. (2019) Structural basis of specific H2A K13/K15 ubiquitination by RNF168, Nat Commun. 10, 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell SJ, Edwards RA, Leung CC, Neculai D, Hodge CD, Dhe-Paganon S & Glover JN (2012) Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation, J Biol Chem. 287, 23900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panier S, Ichijima Y, Fradet-Turcotte A, Leung CC, Kaustov L, Arrowsmith CH & Durocher D. (2012) Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks, Mol Cell. 47, 383–95. [DOI] [PubMed] [Google Scholar]
- 30.Pinato S, Gatti M, Scandiuzzi C, Confalonieri S & Penengo L. (2011) UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway, Mol Cell Biol. 31, 118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi TS, Hirade Y, Toma A, Sato Y, Yamagata A, Goto-Ito S, Tomita A, Nakada S & Fukai S. (2018) Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168, Nat Commun. 9, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Q, Botuyan MV, Cui G, Zhao D & Mer G. (2017) Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18, Mol Cell. 66, 473–487 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munoz MC, Yanez DA & Stark JM (2014) An RNF168 fragment defective for focal accumulation at DNA damage is proficient for inhibition of homologous recombination in BRCA1 deficient cells, Nucleic Acids Res. 42, 7720–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luijsterburg MS, Typas D, Caron MC, Wiegant WW, van den Heuvel D, Boonen RA, Couturier AM, Mullenders LH, Masson JY & van Attikum H. (2017) A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation, Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zong D, Adam S, Wang Y, Sasanuma H, Callen E, Murga M, Day A, Kruhlak MJ, Wong N, Munro M, Ray Chaudhuri A, Karim B, Xia B, Takeda S, Johnson N, Durocher D & Nussenzweig A. (2019) BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation, Mol Cell. 73, 1267–1281 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R & Penengo L. (2015) RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage, Cell Rep. 10, 226–38. [DOI] [PubMed] [Google Scholar]
- 37.Lee JH & Paull TT (2004) Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex, Science. 304, 93–6. [DOI] [PubMed] [Google Scholar]
- 38.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L & Shiloh Y. (2003) Requirement of the MRN complex for ATM activation by DNA damage, EMBO J. 22, 5612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gobbini E, Cesena D, Galbiati A, Lockhart A & Longhese MP (2013) Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks, DNA Repair (Amst). 12, 791–9. [DOI] [PubMed] [Google Scholar]
- 40.Bekker-Jensen S & Mailand N. (2010) Assembly and function of DNA double-strand break repair foci in mammalian cells, DNA Repair (Amst). 9, 1219–28. [DOI] [PubMed] [Google Scholar]
- 41.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S & Mailand N. (2015) Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage, Nature. 527, 389–93. [DOI] [PubMed] [Google Scholar]
- 42.Nowsheen S, Aziz K, Aziz A, Deng M, Qin B, Luo K, Jeganathan KB, Zhang H, Liu T, Yu J, Deng Y, Yuan J, Ding W, van Deursen JM & Lou Z. (2018) L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage, Nat Cell Biol. 20, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, Mailand N, Neumann B, Heriche JK, Shearer R, Saunders D, Bartek J, Lukas J & Lukas C. (2012) TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes, Cell. 150, 697–709. [DOI] [PubMed] [Google Scholar]
- 44.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA & Sixma TK (2012) RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling, Cell. 150, 1182–95. [DOI] [PubMed] [Google Scholar]
- 45.Gatti M, Pinato S, Maspero E, Soffientini P, Polo S & Penengo L. (2012) A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase, Cell Cycle. 11, 2538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F & Durocher D. (2013) 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark, Nature. 499, 50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura K, Sakai W, Kawamoto T, Bree RT, Lowndes NF, Takeda S & Taniguchi Y. (2006) Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks, DNA Repair (Amst). 5, 741–9. [DOI] [PubMed] [Google Scholar]
- 48.Bohgaki M, Bohgaki T, El Ghamrasni S, Srikumar T, Maire G, Panier S, Fradet-Turcotte A, Stewart GS, Raught B, Hakem A & Hakem R. (2013) RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks, Proc Natl Acad Sci U S A. 110, 20982–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guturi KKN, Bohgaki M, Bohgaki T, Srikumar T, Ng D, Kumareswaran R, El Ghamrasni S, Jeon J, Patel P, Eldin MS, Bristow R, Cheung P, Stewart GS, Raught B, Hakem A & Hakem R. (2016) RNF168 and USP10 regulate topoisomerase IIalpha function via opposing effects on its ubiquitylation, Nat Commun. 7, 12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kongsema M, Zona S, Karunarathna U, Cabrera E, Man EP, Yao S, Shibakawa A, Khoo US, Medema RH, Freire R & Lam EW (2016) RNF168 cooperates with RNF8 to mediate FOXM1 ubiquitination and degradation in breast cancer epirubicin treatment, Oncogenesis. 5, e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK & Richard S. (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites, EMBO J. 31, 1865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker JR, Bonnet C, Clifford G, Groth A, Wilson MD & Chapman JR (2020) BARD1 links histone H2A Lysine-15 ubiquitination to initiation of BRCA1-dependent homologous recombination, bioRxiv. [DOI] [PubMed] [Google Scholar]
- 53.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM & Greenberg RA (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites, Science. 316, 1198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Her J, Soo Lee N, Kim Y & Kim H. (2016) Factors forming the BRCA1-A complex orchestrate BRCA1 recruitment to the sites of DNA damage, Acta Biochim Biophys Sin (Shanghai). 48, 658–64. [DOI] [PubMed] [Google Scholar]
- 55.Savage KI & Harkin DP (2015) BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability, FEBS J. 282, 630–46. [DOI] [PubMed] [Google Scholar]
- 56.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E & Livingston DM (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci, Genes Dev. 25, 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coleman KA & Greenberg RA (2011) The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection, J Biol Chem. 286, 13669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lombardi PM, Matunis MJ & Wolberger C. (2017) RAP80, ubiquitin and SUMO in the DNA damage response, J Mol Med (Berl). 95, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwertman P, Bekker-Jensen S & Mailand N. (2016) Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers, Nat Rev Mol Cell Biol. 17, 379–94. [DOI] [PubMed] [Google Scholar]
- 60.Munoz MC, Laulier C, Gunn A, Cheng A, Robbiani DF, Nussenzweig A & Stark JM (2012) RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1, J Biol Chem. 287, 40618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sy SM, Jiang J, Dong SS, Lok GT, Wu J, Cai H, Yeung ES, Huang J, Chen J, Deng Y & Huen MS (2011) Critical roles of ring finger protein RNF8 in replication stress responses, J Biol Chem. 286, 22355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krais JJ, Wang Y, Bernhardy AJ, Clausen E, Miller JA, Cai KQ, Scott CL & Johnson N. (2020) RNF168-Mediated Ubiquitin Signaling Inhibits the Viability of BRCA1-Null Cancers, Cancer Res. 80, 2848–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmid JA, Berti M, Walser F, Raso MC, Schmid F, Krietsch J, Stoy H, Zwicky K, Ursich S, Freire R, Lopes M & Penengo L. (2018) Histone Ubiquitination by the DNA Damage Response Is Required for Efficient DNA Replication in Unperturbed S Phase, Mol Cell. 71, 897–910 e8. [DOI] [PubMed] [Google Scholar]
- 64.Krais JJ & Johnson N. (2020) Ectopic RNF168 expression promotes break-induced replication-like DNA synthesis at stalled replication forks, Nucleic Acids Res. 48, 4298–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weitzman MD, Lilley CE & Chaurushiya MS (2010) Genomes in conflict: maintaining genome integrity during virus infection, Annu Rev Microbiol. 64, 61–81. [DOI] [PubMed] [Google Scholar]
- 66.Weitzman MD, Lilley CE & Chaurushiya MS (2011) Changing the ubiquitin landscape during viral manipulation of the DNA damage response, FEBS Lett. 585, 2897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D & Weitzman MD (2010) A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses, EMBO J. 29, 943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sitz J, Blanchet SA, Gameiro SF, Biquand E, Morgan TM, Galloy M, Dessapt J, Lavoie EG, Blondeau A, Smith BC, Mymryk JS, Moody CA & Fradet-Turcotte A. (2019) Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response, Proc Natl Acad Sci U S A. 116, 19552–19562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujimuro M, Sawada H & Yokosawa H. (1994) Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins, FEBS Lett. 349, 173–80. [DOI] [PubMed] [Google Scholar]
- 70.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP & Citterio E. (2007) Human USP3 is a chromatin modifier required for S phase progression and genome stability, Curr Biol. 17, 1972–7. [DOI] [PubMed] [Google Scholar]
- 71.Lancini C, van den Berk PC, Vissers JH, Gargiulo G, Song JY, Hulsman D, Serresi M, Tanger E, Blom M, Vens C, van Lohuizen M, Jacobs H & Citterio E. (2014) Tight regulation of ubiquitin-mediated DNA damage response by USP3 preserves the functional integrity of hematopoietic stem cells, J Exp Med. 211, 1759–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma N, Zhu Q, Wani G, He J, Wang QE & Wani AA (2014) USP3 counteracts RNF168 via deubiquitinating H2A and gammaH2AX at lysine 13 and 15, Cell Cycle. 13, 106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mosbech A, Lukas C, Bekker-Jensen S & Mailand N. (2013) The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases, J Biol Chem. 288, 16579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uckelmann M, Densham RM, Baas R, Winterwerp HHK, Fish A, Sixma TK & Morris JR (2018) USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A, Nat Commun. 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z, Yang H & Wang H. (2014) The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage, J Biol Chem. 289, 32883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sen Nkwe N, Daou S, Uriarte M, Gagnon J, Iannantuono NV, Barbour H, Yu H, Masclef L, Fernandez E, Zamorano Cuervo N, Mashtalir N, Binan L, Sergeev M, Belanger F, Drobetsky E, Milot E, Wurtele H, Costantino S & Affar EB (2020) A potent nuclear export mechanism imposes USP16 cytoplasmic localization during interphase, J Cell Sci. 133. [DOI] [PubMed] [Google Scholar]
- 77.Typas D, Luijsterburg MS, Wiegant WW, Diakatou M, Helfricht A, Thijssen PE, van den Broek B, Mullenders LH & van Attikum H. (2016) The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80, Nucleic Acids Res. 44, 2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH, Ordog T, Lou Z, You Z & Zhang Z. (2016) USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response, Genes Dev. 30, 946–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu M, Liu K, Mao Z, Luo J, Gu W & Zhao W. (2016) USP11 Is a Negative Regulator to gammaH2AX Ubiquitylation by RNF8/RNF168, J Biol Chem. 291, 959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartocci C & Denchi EL (2013) Put a RING on it: regulation and inhibition of RNF8 and RNF168 RING finger E3 ligases at DNA damage sites, Front Genet. 4, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swaney DL, Rodriguez-Mias RA & Villen J. (2015) Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover, EMBO Rep. 16, 1131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D & Gygi SP (2003) A proteomics approach to understanding protein ubiquitination, Nat Biotechnol. 21, 921–6. [DOI] [PubMed] [Google Scholar]
- 83.Walser F, Mulder MPC, Bragantini B, Burger S, Gubser T, Gatti M, Botuyan MV, Villa A, Altmeyer M, Neri D, Ovaa H, Mer G & Penengo L. (2020) Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response, Mol Cell. 80, 423–436 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alatwi HE & Downs JA (2015) Removal of H2A.Z by INO80 promotes homologous recombination, EMBO Rep. 16, 986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y & Price BD (2012) Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair, Mol Cell. 48, 723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O & Oberdoerffer P. (2014) A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance, Cell Rep. 8, 1049–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sebastian R, Hosogane EK, Sun EG, Tran AD, Reinhold WC, Burkett S, Sturgill DM, Gudla PR, Pommier Y, Aladjem MI & Oberdoerffer P. (2020) Epigenetic Regulation of DNA Repair Pathway Choice by MacroH2A1 Splice Variants Ensures Genome Stability, Mol Cell. 79, 836–845 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J, Sturgill D, Sebastian R, Khurana S, Tran AD, Edwards GB, Kruswick A, Burkett S, Hosogane EK, Hannon WW, Weyemi U, Bonner WM, Luger K & Oberdoerffer P. (2018) Replication Stress Shapes a Protective Chromatin Environment across Fragile Genomic Regions, Mol Cell. 69, 36–47 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leung JW, Agarwal P, Canny MD, Gong F, Robison AD, Finkelstein IJ, Durocher D & Miller KM (2014) Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling, PLoS Genet. 10, e1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei Y, Wang HT, Zhai Y, Russell P & Du LL (2014) Mdb1, a fission yeast homolog of human MDC1, modulates DNA damage response and mitotic spindle function, PLoS One. 9, e97028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chahwan R, Gravel S, Matsusaka T & Jackson SP (2013) Dma/RNF8 proteins are evolutionarily conserved E3 ubiquitin ligases that target septins, Cell Cycle. 12, 1000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ & Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks, Cell. 123, 1213–26. [DOI] [PubMed] [Google Scholar]
- 93.Chroma K, Mistrik M, Moudry P, Gursky J, Liptay M, Strauss R, Skrott Z, Vrtel R, Bartkova J, Kramara J & Bartek J. (2017) Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress, Oncogene. 36, 2405–2422. [DOI] [PubMed] [Google Scholar]