Abstract
The molecular weight and isoelectric point (pI) of the proteins plays important role in the cell. Depending upon the shape, size, and charge, protein provides its functional role in different parts of the cell. Therefore, understanding to the knowledge of their molecular weight and charges is (pI) is very important. Therefore, we conducted a proteome-wide analysis of protein sequences of 689 fungal species (7.15 million protein sequences) and construct a virtual 2-D map of the fungal proteome. The analysis of the constructed map revealed the presence of a bimodal distribution of fungal proteomes. The molecular mass of individual fungal proteins ranged from 0.202 to 2546.166 kDa and the predicted isoelectric point (pI) ranged from 1.85 to 13.759 while average molecular weight of fungal proteome was 50.98 kDa. A non-ribosomal peptide synthase (RFU80400.1) found in Trichoderma arundinaceum was identified as the largest protein in the fungal kingdom. The collective fungal proteome is dominated by the presence of acidic rather than basic pI proteins and Leu is the most abundant amino acid while Cys is the least abundant amino acid. Aspergillus ustus encodes the highest percentage (76.62%) of acidic pI proteins while Nosema ceranae was found to encode the highest percentage (66.15%) of basic pI proteins. Selenocysteine and pyrrolysine amino acids were not found in any of the analysed fungal proteomes. Although the molecular weight and pI of the protein are of enormous important to understand their functional roles, the amino acid compositions of the fungal protein will enable us to understand the synonymous codon usage in the fungal kingdom. The small peptides identified during the study can provide additional biotechnological implication.
Subject terms: Computational biology and bioinformatics, Microbiology, Molecular biology, Systems biology, Biochemistry, Proteins, Proteomics
Introduction
Genomic studies have contributed enormously towards understanding the molecular mechanisms underlying cellular development through the expression of proteins, collectively referred to as the proteome1,2. Proteins play a qualitative and quantitative role in growth, development, and stress tolerance of an organism3–6. They are the structural and functional units of a cell comprising a chain of amino acids which are arranged as determined by their coding sequences (mRNA)7–9. Different proteins are synthesized based on the immediate and developmental needs required by the cell as part of programmed development or specific response signals10,11. Proteins often undergo post-translational modification and are frequently targeted for delivery to specific sub-cellular locations12,13. The function of a protein depends upon its molecular structure and is also affected by the post-translational modifications that occur14–17. Additionally, the sub-cellular localization of a protein plays a critical role in defining its function since different cellular compartments regulate various physiological and biochemical functions18–20. Similarly, post-translational modifications also play multifunctional regulatory roles that change the function of a protein and regulate biological processes21–23. The pH of the cytoplasm plays an important role for the post-translational modification that occur and directing the sub-cellular localization of proteins24–27. Tight regulation of proton fluxes and the maintenance of a pH gradient across cellular membranes play critical roles in cellular homeostasis24,28–30. Different proteins respond to diverse pH gradients across cellular compartments that regulate cell metabolism31,32. Large deviations in pH, however, need to be avoided to maintain normal metabolic and intracellular biochemical processes. The isoelectric point (pI) of a protein molecule plays a major role in its contribution to cellular homeostasis. The pI is the pH at which the protein molecule carries no electrical charges. The net charge of a protein molecule is affected by the pH gradient of a cell and by sub-cellular compartments. This can lead to a more positive or negatively charged protein molecule and determine the gain or loss of protons (H+).
Although protein synthesis occurs in the cytoplasm (pH 7.4–7.7)33, proteins are often transported to different sub-cellular locations, including the endoplasmic reticulum, Golgi complexes, mitochondria, vacuoles (pH 6.2), cell membranes, lysosomes, and other compartments after synthesis and processing34. Therefore, different protein needs to undergo modifications that are appropriate to the pH gradient that exists across different cellular compartments. The pH of the cytoplasm is slightly alkaline35,36 while the pH of sub-cellular organelles, including the vacuole and lysosomes, are slightly acidic34. The pI of a protein has been used as a standard parameter to distinguish proteins37. The molecular mass of a protein also plays a vital role in its translocation across the cellular compartments38,39. Therefore, knowing the pI and molecular mass of a protein provides critical information that can help to determine its functional role in a cell. The advent of next-generation, high-throughput DNA sequencing allowed for the generation of complete genome sequences of more than one thousand fungal species. In the current study, the annotated protein sequences of 689 species derived from completed genome sequencing efforts within public repositories were used to construct a collective 2-D map of the fungal proteome.
Results and discussion
Size of the fungal genome is not directly proportional to the number of amino acids
A proteome-wide study of 689 fungal species was conducted to characterize the molecular mass, isoelectric point, and amino acid composition of the fungal kingdom. The study comprised 7.159 million protein sequences from ten fungal groups, Ascomycota (466 species), Basidiomycota (148 species), Blastocladiomycota (1 species), Chytridiomycota (8 species), Glomeromycota (1 species), Microsporidia (26 species), Mucoromycota (23 species), Neocallimastigomycota (1 species), Opisthokonta (1 species), and Zoopagomycota (14 species). Results indicated that, Fibula rzhizoctina (Basidiomycota), commonly known as the cuckoo fungus, encoded the highest number protein sequences, totalling 32,854 (Supplementary File 1). The largest genome size in the fungal kingdom was found in Neocallimastix californiae (193.032 Mb) followed by Tuber magnatum (192.781 Mb) (Supplementary File 2). Although Neocallimastix californiae (193.032 Mb) had the largest genome, Fibula rhizoctonia (genome size 95.118 Mb) encoded a higher number of amino acids (10.940068 million) (Supplementary Fig. 1A, Supplementary File 1). The genome size of all 689 fungal species was compared with the number of amino acids using regression analysis to determine the correlation between genome size and the number of amino acids in fungi (Supplementary Fig. 2A,B). Results indicated that genome size in the fungal kingdom is not proportional to the number of amino acids encoded by a genome (Supplementary Fig. 2A). This indicates that the larger fungal genomes contain a greater number of introns and other non-coding nucleotide sequences in their genomes. A linear regression analysis of the presence of a specific amino acid number in a fungal species revealed that Met (0.993) had the highest correlation coefficient, followed by Thr (0.9917), Leu (0.9913), Trp (0.9907), and Val (0.9906). The lowest correlation coefficient was found for Asn (0.8828) (Supplementary Fig. 2B).
The average molecular mass of fungal protein was 50.96 kDa and fungal proteome was 518539.109524477 kDa
The average molecular mass of fungal proteins was 50.96 kDa (Supplementary File 3). The highest average molecular mass of proteins was found in Aspergillus ustus (70.7 kDa), followed by Ascosphaera apis (69.13 kDa), Candida boidinii (66.71 kDa), Moesziomyces antacticus (66.37 kDa), and Anthracocystis floculosa (66.21 kDa) (Supplementary File 3). The lowest average molecular mass of proteins found in Moniliopthora perniciosa (20.34 kDa), Aspergillus wentii (26.94), and Hepatospora eriocheir (26.94 kDa) (Supplementary File 3). Approximately 9.29% of fungal proteins have a mass > 100 kDa. When the molecular mass of the whole proteome of individual species was analysed, Rhizophgus clarus was found to have the heaviest proteome with a total molecular mass of 1,210,411.214 kDa, followed by Fibularhizoctina sp. (1,204,483.772 kDa), Diversispora versiformis (1,171,535.352 kDa), and Neocallimatrix californiae (1,120,727.893 kDa). The average molecular mass of a fungal proteome was found to be 518,539.109 kDa (Supplementary File 3). Further analysis revealed that the largest protein in the fungal kingdom was a non-ribosomal peptide synthase (accession number RFU80400.1) found in Trichoderma arundinaceum. This protein has a molecular mass of 2546.166 kDa and is the first fungal protein ever reported with highest molecular mass. Other high molecular mass proteins include an amino acid adenylation protein (2542.314 kDa, 23,089 aa, accession number PTB76898.1) in Trichoderma longibrachiatum, NRPS protein (2527.788 kDa, 22,960 aa, accession number OTA01063.1) in Trichoderma parareesei, and a non-ribosomal peptide synthase (2314.108 kDa, 20,891 aa, accession number EHK18913.1) in Trichoderma virens. The largest protein molecules are act as molecular machines of the cell that provides the structural and functional unit to the cell40. Non-ribosomal peptide synthase is a multi-domain protein that produces several important secondary metabolites for its virulence41. Fungal NRPS protein comprises of several pharmacological relevant compounds including β-lactam anti-biotics, echinocandin antibiotics, and cyclosporins41.
The molecular mass of the smallest fungal protein/peptide was 0.202 kDa (hypothetical protein, accession number KFA68322.1) found in Stachybotrys chlorohalonata. It is a dipeptide with A-L amino acids. Some other low molecular mass dipeptides were M-G (0.206 kDa, accession number OJA20730.1) found in Rhizopogon vesiculosus, R-G (0.231 kDa, accession number RDB21682.1) found in Hypsizygus marmoreus, and M-V (0.248 kDa, accession number SCN86923.1) found in Fusarium fujikuroi. A low molecular weight tripeptide was A-L-K (0.303 kDa, accession number KUM56815.1) and tetrapeptide was M-G-G-T (0.364 kDa, accession number PNY18451.1). The present study is the first to identify A-L, M-G, and R-G dipeptides; A-L-K tripeptides; and M-G-G-T tetrapeptides in the fungal kingdom. These small peptides can function as novel bioactive molecules that regulate several biological processes42–44. Small peptides can act as a hormone45,46, biocide47, and as an anti-cancer therapeutic agent48,49. The presence of these small peptides and their activity can have a significant impact on the physiology of an organism. Therefore, the function of these small compounds should be further explored for their potential application and commercialization.
Isoelectric points (pI) of the fungal proteome ranges from 1.85 to 13.759
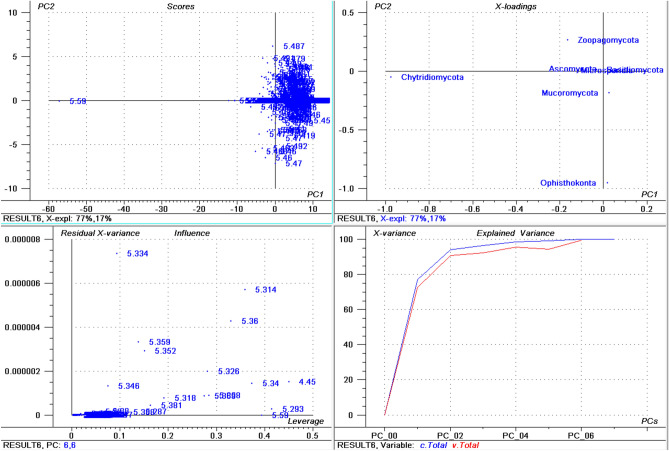
The isoelectric point of proteins in the collective fungal proteome ranged from 1.85 in Candida maltose (interspersed repeat antigen (FIRA) protein, partial; accession: EMG49333.1) to 13.759 in Ophiocordyceps sinensis (hypothetical protein, accession: EQK98935.1). The average acidic pI was 5.222 while the average basic pI was found to be 8.489 (Supplementary File 3, Supplementary Fig. 1B,C). Among a total of 22 identified proteinogenic amino acids, the protein with the highest pI (EQK98935.1) was composed of only ten amino acids, Ala (63), Gly (42), Lys (21), Leu (43), Met (1), Gln (21), Arg (84), Ser (65), Val (42), and Trp (22). The Cys amino acid which is responsible for the formation of disulphide bonds in a protein was not present in the high pI protein. In addition, the negatively charged amino acids Asp and Glu were also not a part of the protein. Approximately 61.68% of the fungal proteins had an acidic pI while only 38.04% of the proteins had a basic pI, indicating that acidic pI proteins were more abundant than the basic pI proteins. The average of highest pI protein was found to be 12.446 (Supplementary Fig. 1D). When proteins with the highest pI in each phylum were analysed, the highest average pI was found in Mucoromycota (12.575) followed by Zoopagomycota (12.548), Basidiomycota (12.542), Chytridiomycota (12.497), Ascomycota (12.431), and Microsporidia (11.937). None of the species within the Microsporidia were found to encode a protein with a pI greater than 12.544. The average lowest pI among fungal phyla was found to be 2.929 (Supplementary Fig. 1E), with the lowest pI found in Zoopagomycota (2.441) followed by Chytridiomycota (2.64), Mucoromycota (2.749), Basidiomycota (2.897), Ascomycota (2.954), and Microsporidia (3.28). Aspergillus ustus was found to encode the highest percentage (76.62%) of acidic pI proteins, followed by Eutypa lata (74.58%), Talaromyces amestolkiae (74.16%), and Pichia membranifaciens (73.84%) (Supplementary File 3). Aspergillus ustus is a microfungus associated with human nail infections and Eutypa lata causes Eutypa dieback disease in grape (Vitis vinifera)50. These species belong to the phylum Ascomycota. The highest percentage of acidic pI proteins in the phylum Basidiomycota was found in Malassezia vespertilionis (68.16%) and the lowest percentage of acidic pI proteins was found in Terfezia bouderi (20.18%) followed by Nosema ceranae (33.49%), and Puccinia sorghi (33.90%) (Supplementary File 3). Among the 689 species, only 40 species (5.80%) contained basic pI proteins indicating that 94.19% of fungal species contain ≥ 50% acidic pI proteins in their proteome. A principal component analysis (PCA) of acidic pI proteins revealed that Ascomycota, Basidiomycota, and Microsporidia cluster together while the Zoopagomycota, Chytridiomycota, Mucoromycota, and Ophisthokonta cluster separately (Fig. 1); suggesting that the percentages of acidic pI protein contribute to the clustering.
Figure 1.
Principal component analysis (PCA) of acidic pI proteins in the fungal proteome. The figure illustrates the relationship between the acidic pI of Ascomycota, Basidiomycota, Microsporidia, Zoopagomycota, Chytridiomycota, and Opthisthokonta. The acidic pI of Chytridiomycota and Opthisthokonta cluster independently from the other phyla. Within the figure (a) scores: reflects the similarities in sample grouping, (b) loading: represents the relative position of a variable and how it relates a sample to different variables, (c) influence plot: represents the Q- or F-residuals vs Leverage or Hotelling T2 statistics that shows the residual statistics on the ordinate axis of sample distance to model and (d) variance: represent the variation in the data of different components. Total residual variance is calculated as a sum of the square of residuals for all the variables, divided by the number of degrees of freedom. The green colour indicates the calibration and the red indicates the validation.
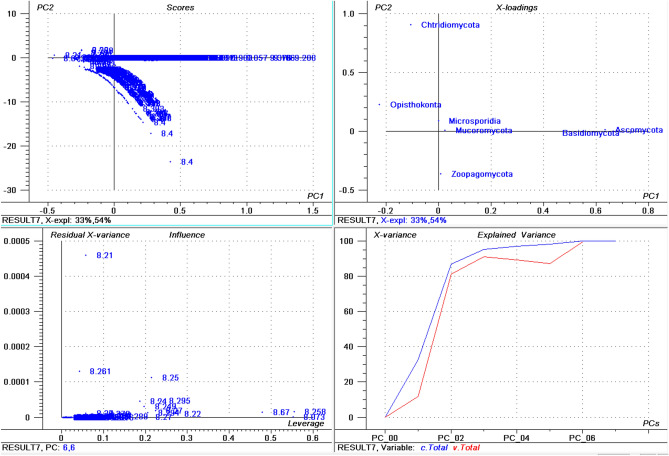
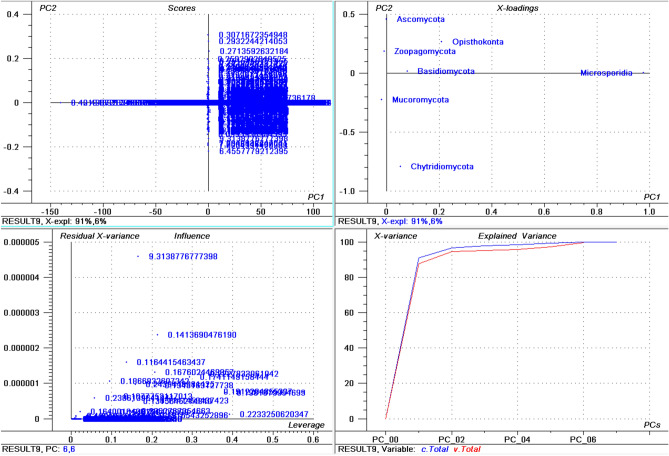
The highest percentage of basic pI proteins was found in Nosema ceranae (66.15%) followed by Puccinia sorghi (65.89%), and Tubulinosema ratisbonensis (63.55%) (Supplementary File 1). Nosema ceranae is the causal agent of nosemosis (a disease in honey bees) and its spores are resistant to high temperature and dehydration. This fungus also produces an insecticide, similar to fipronil/nicotinoids, which causes male sterility in honey bees51,52. The lowest percentage of basic pI proteins was found in Aspergillus ustus (23.22%), followed by Eutypa lata (25.26%), and Fonsecaea erecta (25.59%) (Supplementary File 3). The highest percentage (≥ 60%) of basic pI proteins was found in species within the Microsporidia and Basidiomycota group. In contrast, none of the species in the Ascomycota was found to encode ≥ 60% basic pI proteins in it. The PCA of basic pI proteins revealed that Microsporidia and Mucoromycota cluster together while Ascomycota and Basidiomycota cluster together (Fig. 2). However, Chytridiomycota, Ophisthokonta, and Zoopagomycota cluster independently (Fig. 2). This suggests that the basic pI proteins in the Microsporidia with Mucoromycota and Ascomycota with Basidiomycota share a common pattern of basic pI. Notably, the PCA of the percentage of neutral pI proteins did not exhibit any distinct clustering of phyla (Fig. 3). All of the groups appeared to locate independently from each other, suggesting a great diversity in the percentage of neutral pI proteins in the fungal proteome.
Figure 2.
Principal component analysis (PCA) of basic pI proteins in the fungal proteome. The figure illustrates the relationship between the basic pI of Ascomycota, Basidiomycota, Microsporidia, Zoopagomycota, Chytridiomycota, and Opthisthokonta. The basic pI of Chytridiomycota, Opthisthokonta, and Zoopagomyota cluster independently from the other fungal phyla. In the figure (a) scores: reflects the similarities in sample grouping, (b) loading: represents the relative position of a variable and how it relates a sample to different variables, (c) influence plot: represents the Q- or F-residuals vs Leverage or Hotelling T2 statistics that shows the residual statistics on the ordinate axis of sample distance to model, and (d) variance: represent the variation in the data by different components. Total residual variance is calculated as a sum of square of residuals for all the variables, divided by the number of degrees of freedom. The green color indicates the calibration and the red indicates the validation. The collective fungal proteome was found to contain an average of 17.449 (0.172%) neutral pI proteins (Fig. 1F). The pI of the entire plant kingdom has been reported to range from 1.99 to 13.9642. The lowest pI found for fungal proteins was less than the lowest plant protein pI. In contrast, the highest pI found for a fungal protein was lower than the highest pI found for a plant protein.
Figure 3.
Principal component analysis (PCA) of percentage (%) of occurrence of neutral pI proteins in the fungal proteome. The figure illustrates the relationship between the percentage of neutral pI proteins in the Ascomycota, Basidiomycota, Microsporidia, Zoopagomycota, Chytridiomycota, and Opthisthokonta.
The Mol. Weight and pI of fungal proteins exhibit a bimodal distribution
The pI of the analysed fungal proteins, ranging from 1.85 to 13.759 and exhibited a bimodal distribution (Fig. 4). The bimodal distribution of the pI and molecular mass of the fungal proteome provides a virtual 2-D proteome map of the fungi (Fig. 4). Mohanta et al., (2019) previously reported a trimodal distribution of plant proteomes42. Schwartz et al., also reported a trimodal distribution of the pI of eukaryotic proteins37. The pH of cytoplasm is close to neutral while the majority of fungal proteins fall in the acidic pI range (Fig. 4). Approximately 94.21% of the fungal species encode proteins whose proteome contain more than 50% of acidic pI proteins. The pI peak of acidic proteins was more prominent compared to basic and neutral proteins (Fig. 4). Kiraga et al., (2007) reported that acidic and basic pI properties of the proteins are the basis of bi- and trimodal distribution53. They also reported that taxonomy, ecological niche, and sub-cellular localization play an important role in determining the acidity and basicity of a protein53. Therefore, we analysed the normal probability distribution of the percentage of all of the acidic pI proteins from all fungal phyla (Ascomycota, Basidiomycota, Chytridiomycota, Microsporidia, Mucoromycota, and Zoopagomycota). Results indicated that the correlation coefficient (0.9635) tended slightly towards linearity, irrespective of taxonomical hierarchy (Supplementary Fig. 3A). Similarly, the normal probability distribution of the percentage of basic pI proteins in all taxonomical groups of fungi was calculated. Results indicated that the correlation coefficient was 0.974 (Supplementary Fig. 3B). Basic pI proteins were correlated in a greater extent with their number and percentage in the collective fungal proteome compared to the acidic pI proteins. No correlation was found between acidic or basic pI proteins and their taxonomy. Knight et al.54 also reported a negative correlation between the pI and taxonomy of an organism as well as a negative correlation between the pI of proteins and the phylogeny of an organism54. Only 0.172% of the proteins in fungi were found in the neutral pI range (Supplementary File 3). Tilletia controversa had the highest percentage (2.231%) of neutral pI proteins and Anncaliia algerae had the lowest percentage (0.027%) of neutral proteins (Supplementary File 3).
Figure 4.
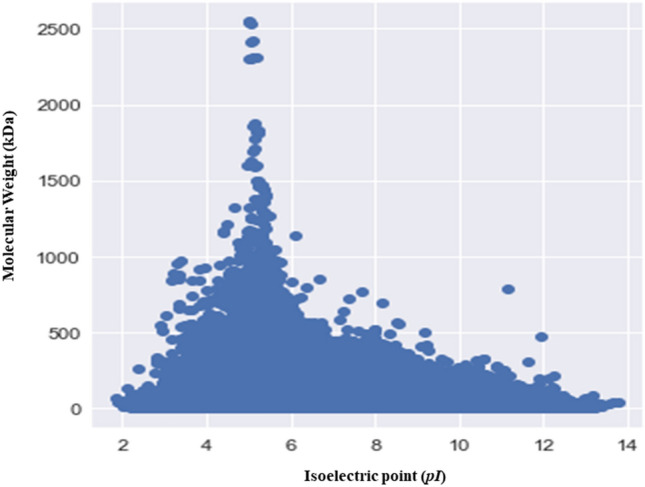
Virtual 2D map of the fungal proteome. The virtual map reveals the bimodal distribution of molecular mass (kDa) and isoelectric point (pI) in the collective fungal proteome. The X-axis represents the isoelectric point and Y-axis represents the molecular mass of the fungal proteins.
The provided molecular mass and pI of the fungal proteomes are based on the annotated protein sequences. During the event of protein translation, the protein sequences undergo post-translational modification including methylation, phosphorylation, acetylation, hydroxylation, amidation, ubiquitylation, sulfation, glycosylation, and other55,56. These post-translational modification event changes the charges of the protein molecule57. Therefore, the predicted pI of the protein may be little different from the exact pI. Kumar et al., (2004) reported that the extent of shift in pI of a protein upon phosphorylation is dependent on its size and native pI58. Therefore, the native pI of a protein plays critical role as well. The shift of pI of a protein in a particular posttranslational event also vary greatly58. A protein with lower molecular weight tend to show pI-shift more frequently than the higher molecular weight proteins58. Addition of at least phosphorylation to a protein with a molecular mass of 110 kDa result in a pI-shift of less than 0.2 units and an addition of the same amount of phosphorylation with a molecular weight of 11.8 kDa result in a pI-shift of approximately 1 unit58. Protein with basic pI show maximum pI-shift upon phosphorylation while acidic pI protein shows a relatively lower pI-shift58. The fungal proteome is predominated by acidic pI proteins compared to the basic pI proteins. Therefore, the pI-shift of fungal acidic pI proteome will be relatively lower. However, phosphorylation and dephosphorylation of a protein is a continuous process and none of a particular protein ever stay in either at phosphorylation or dephosphorylation state. Therefore, the pI-shift due to change in phosphorylation also revert back to the original state upon dephosphorylation. Cathepsin B protein upon single phosphorylation shifts its pI from 5.23 to 5.15 (experimental)59 and heat induced nuclear accumulation protein HSC70 upon single phosphorylation shifts it pI from 5.48 to 5.43 (experimental)60.
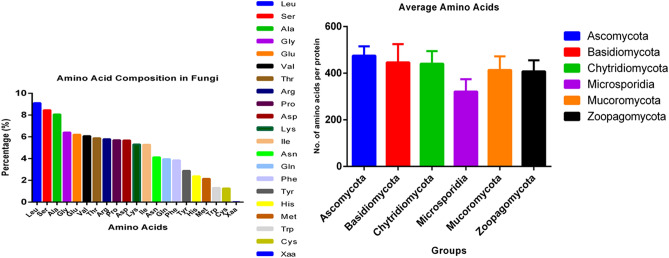
Leu is a high-abundant and Cys is a low-abundant amino acid in the Fungal Kingdom
The amino acid composition of proteins in the fungal kingdom was analysed to determine which amino acids were more abundant and which were lower in abundance (Supplementary File 1). It was estimated that fungi encoded 4.675520004 million amino acids per proteome. The proteome of Fibula rhizoctonia encoded highest number of amino acids (10.940068 million) followed by Sphaerobolus stellatus (10.818606 million), Rhizophagus clarus (10.524515 million), and Diversispora versiformis (10.170037 million) (Supplementary File 2). The analysis revealed that Leu (9.115%) was the most abundant amino acid in the collective fungal proteome while Cys (1.267%) was the least abundant (Table 1). The high abundance of Leu was followed by Ser (8.465%), Ala (8.066%) and Gly (6.41%); while the low abundance of Cys (1.267%) was followed by Trp (1.322%) (Table 1). As Leu, Ala, and Gly are non-polar amino acids, it seems the fungal proteome favours the synthesis of non-polar amino acids than other types. It would be very interesting to determine the reason behind the differential abundance of amino acids in the fungal proteome. When the half-life of individual amino acids was compared, Leu was found to have the shortest half-life (namely, 3 min), while the half-life of Cys was more than 20 h61–64. Since Leu has very short half-life period, it needs to encode a greater number of Leu amino acid in the cell to maintain protein synthesis machinery. Cys has a very long half-life and thus the degradation of Cys in the cell is relatively slower than Leu. Therefore, it seems that fungal cells synthesize a lower number of Cys amino acids. Cys amino acid used to produce glutathione anti-oxidant as well as amino acid taurine. Cys also sometimes converted to glucose for the source of energy65. A cross-talk between Cys and glutathione is critical for the regulation of amino acid signaling pathway66. The role of Cys as the precursor molecule in plant is also highly important other than its role in the protein67. Its role in plant stress response is highly important for its redox activity67. Accumulation of Pro amino acid occurs due to osmotic stress in plants and it act as a plant abiotic stress marker68. However, accumulation of other amino acids like Asp, Glu, Ser, Gly, and Gln also occur due to biotic and abiotic stress responses69.
Table 1.
Amino acid composition of the fungal proteomes.
| Amino acids | Composition (%) |
|---|---|
| Leu | 9.115056391 |
| Ser | 8.465427891 |
| Ala | 8.066886639 |
| Gly | 6.416593956 |
| Glu | 6.212047906 |
| Val | 6.080098604 |
| Thr | 5.885447967 |
| Arg | 5.79174785 |
| Pro | 5.70190633 |
| Asp | 5.677441609 |
| Lys | 5.309656746 |
| Ile | 5.291616955 |
| Asn | 4.128360538 |
| Gln | 3.960659614 |
| Phe | 3.856287715 |
| Tyr | 2.889458344 |
| His | 2.388615762 |
| Met | 2.156579159 |
| Trp | 1.32229263 |
| Cys | 1.267849975 |
| Xaa | 0.015967419 |
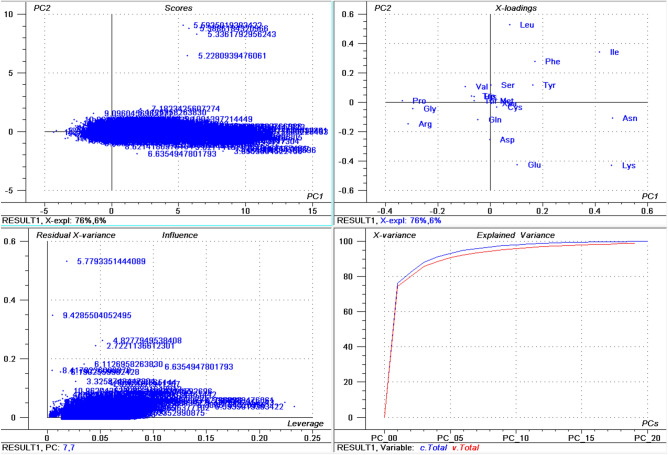
PCA analysis of the amino acid composition of fungal proteins revealed that Cys, Met, Xaa (unknown amino acids), Thr, Trp, Val, and Ser clustered together while Leu, Ile, Phe, Asn, Lys, Glu, and Ala locate separately (Fig. 5). Further analysis indicated that the highest percentage of Ala (14.00%) and Arg (7.76%) was found in Tilletiopsis washingtonensis and the highest percentage of Cys (2.40%), Met (3.78%), and Val (7.29%) was found in Ordospora colligate (Table 2). The lowest percentage of Phe (2.842%), Ile (3.039%), Lys (3.359%), and Asn (2.139%) was found in Tilletiopsis washingtonensis, while the lowest percentage of Cys (0.533%), Asp (3.023%), Gln (1.956%), and Arg (2.383%) was found in Trichoderma asperellum (Table 2).
Figure 5.
PCA analysis of amino acid composition in fungal proteomes. Analysis revealed that Pro, Gly, and Arg; Val, Ser, Met, Cys, Gln, Trp, Thr, His, and Ala cluster together while Leu, Ile, Phe, Tyr, Asn, Asp, Glu, and Lys group independently from the other amino acids.
Table 2.
Highest and lowest composition of amino acids in the fungal kingdom.
| Amino acids | Highest percentage (%) | Name of the species with highest abundance | Lowest percentage (%) | Name of the species with lowest abundance | Average percentage (%) | Standard deviation | variance |
|---|---|---|---|---|---|---|---|
| Ala | 14.009 | Tilletiopsis washingtonensis | 2.315 | Nosema apis | 8.066 | 1.678 | 0.593 |
| Cys | 2.407 | Ordospora colligata | 0.533 | Trichoderma asperellum | 1.267 | 0.193 | 0.039 |
| Asp | 6.428 | Pichia membranifaciens | 3.023 | Trichoderma asperellum | 5.677 | 0.302 | 0.016 |
| Glu | 9.651 | Amphiamblys sp | 2.836 | Penicillium roqueforti | 6.212 | 0.519 | 0.203 |
| Phe | 7.845 | Penicillium nordicum | 2.84 | Tilletiopsis washingtonensis | 3.856 | 0.535 | 0.396 |
| Gly | 8.035 | Jaminaea rosea | 3.00 | Edhazardia aedis | 6.416 | 0.87 | 0.073 |
| His | 3.300 | Malassezia restricta | 1.471 | Hepatospora eriocheir | 2.388 | 0.233 | 0.025 |
| Ile | 11.170 | Trichoderma asperellum | 3.039 | Tilletiopsis washingtonensis | 5.291 | 1.174 | 1.779 |
| Lys | 11.063 | Tubulinosema ratisbonensis | 3.359 | Tilletiopsis washingtonensis | 5.309 | 1.293 | 1.920 |
| Leu | 14.101 | Penicillium roqueforti | 7.753 | Piromyces sp. | 9.115 | 0.523 | 0.351 |
| Met | 3.787 | Ordospora colligata | 1.462 | Ascoidea rubescens | 2.156 | 0.189 | 0.032 |
| Asn | 10.944 | Anaeromyces robustus | 2.139 | Tilletiopsis washingtonensis | 4.128 | 1.29 | 2.366 |
| Pro | 7.272 | Cutaneotrichosporon oleaginosum | 2.00 | Enterocytozoon hepatopenaei | 5.701 | 0.954 | 0.071 |
| Gln | 5.278 | Absidia repens | 1.956 | Trichoderma asperellum | 3.960 | 0.37 | 0.050 |
| Arg | 7.766 | Tilletiopsis washingtonensis | 2.383 | Trichoderma asperellum | 5.791 | 0.893 | 0.078 |
| Ser | 11.474 | Smittium mucronatum | 6.322 | Enterocytozoon bieneusi | 8.465 | 0.61 | 0.233 |
| Thr | 6.776 | Absidia glauca | 4.057 | Encephalitozoon cuniculi | 5.885 | 0.332 | 0.014 |
| Val | 7.295 | Ordospora colligata | 4.315 | Ascoidea rubescens | 6.080 | 0.407 | 0.035 |
| Trp | 1.647 | Nectria haematococca | 0.431 | Edhazardia aedis | 1.322 | 0.227 | 0.001 |
| Tyr | 5.081 | Spraguea lophii | 0.053 | Candida orthopsilosis | 2.889 | 0.495 | 0.261 |
| Xaa | 1.719 | Rhizoctonia solani | 0.000 | 0.015 | 0.115 |
The lowest percentage of Gly (3.00) and Trp (0.431) was found in Edhazardia aedis, while the lowest percentage of Met (1.462%) and Val (4.315%) was found in Ascoidea rubescens (Table 2). Amino acid abundance was also examined by grouping species into different phyla. Results indicated that amino acid abundance tended to be phyla-specific (Table 3).
Table 3.
Distribution of average amino acid composition (%) in different groups of the fungal kingdom.
| Amino acids | Ascomycota | Basidiomycota | Microsporidia | Chytridiomycota | Mucoromycota | Zoopagomycota |
|---|---|---|---|---|---|---|
| Ala | 8.1 | 9.06 | 4.37 | 6.82 | 6.63 | 6.83 |
| Cys | 1.23 | 1.22 | 1.89 | 1.44 | 1.41 | 1.33 |
| Asp | 5.72 | 5.58 | 5.39 | 5.59 | 5.79 | 5.52 |
| Glu | 6.22 | 5.92 | 7.54 | 6.24 | 6.22 | 6.08 |
| Phe | 3.86 | 3.54 | 5.18 | 3.87 | 3.98 | 4.05 |
| Gly | 6.54 | 6.66 | 4.62 | 5.56 | 5.18 | 5.72 |
| His | 2.35 | 2.56 | 1.93 | 2.21 | 2.64 | 2.22 |
| Ile | 5.24 | 4.62 | 8.27 | 6.32 | 6.06 | 6.13 |
| Lys | 5.26 | 4.5 | 8.85 | 6.44 | 6.19 | 6.24 |
| Leu | 9.07 | 9.19 | 9.53 | 8.78 | 9.12 | 9.18 |
| Met | 2.15 | 2.07 | 2.46 | 2.16 | 2.39 | 2.18 |
| Asn | 4.11 | 3.34 | 6.43 | 6.08 | 5 | 5.51 |
| Pro | 5.68 | 6.46 | 3.05 | 5 | 4.97 | 4.86 |
| Gln | 4.02 | 3.87 | 2.88 | 3.76 | 4.62 | 3.93 |
| Arg | 5.76 | 6.34 | 4.56 | 4.75 | 5.06 | 4.99 |
| Ser | 8.37 | 8.88 | 7.45 | 8.58 | 8.28 | 9.2 |
| Thr | 5.91 | 5.95 | 4.97 | 5.97 | 6.06 | 5.75 |
| Val | 6.08 | 6.19 | 5.8 | 5.96 | 5.75 | 5.86 |
| Trp | 1.35 | 1.39 | 0.66 | 1.08 | 1.19 | 1.04 |
| Tyr | 2.88 | 2.54 | 4.01 | 3.3 | 3.36 | 3.28 |
Fungi do not encode selenocysteine (Sec) and pyrrolysine (Pyl) amino acids
Selenocysteine (Sec) and Pyrrolysine (Pyl) are considered proteinogenic amino acids. These amino acids were found to be absent in any of the 7.15 million fungal protein sequences examined so far. A selenoproteomic study in yeast also did not find the presence of Sec70. The presence of some unknown amino acids (Xaa) was also found in our analysis (Supplementary File 1). On average, 0.015% (statistically insignificant) of fungal proteins contain a Xaa amino acid. The highest percentage (1.719%) of Xaa amino acids was found in Rhizoctonia solani followed, by Brettanomyces bruxellensis (1.64%), and Trachipleistophora hominis (1.19%) (Supplementary File 1). Among the 689 examined species, 317 contained Xaa amino acids. In addition to Xaa amino acids, a few species also encoded ambiguous amino acids B, J, and Z amino acids (Supplementary File 1). The highest number of B (1415), J (363), and Z (405) amino acids was found in Brettanomyces bruxellensis, Hanseniaspora opuntiae, and Hanseniaspora uvarum, respectively (Supplementary File 1). The ambiguous amino acid B stands for either Asn or Asp and translates into Asn; J stands for Leu or Ile and translate into Leu; and Z stands for Gln or Glu and translates into Gln. Previous studies by Mariotti et al., (2019) and Jiang et al., (2012) also reported the absence of Sec in fungal proteins71,72. Mariotti et al., (2019), however, have reported that fungi do contain Sec amino acids73. They reported the presence of Sec amino acids encoding machinery in nine species Bifiguratus adelaide, Gonapodya prolifera, Capniomyces stellatus, Zancudomyces culisetae, Smittium angustum, and Furculomyces boomerangus73. This stands in contrast to the findings of the present study for its translated product. To further validate the findings of the present study, we re-analysed the proteome sequences of all the nine species mentioned by Mariotti et al., (2019) but did not find the presence of any Sec (U) amino acids (Supplementary Fig. 4). Mariotti et al., (2019) utilized a BLASTN approach in their study, which might be the cause of the noted contradiction73. When we searched for Sec-encoding protein sequences in the 7.15 million fungal proteome sequences from 689 species, we found that 134 sequences in 112 fungal species had amino acid sequences with an annotation name “selenocysteine” (Supplementary File 4). When we downloaded and searched 134 protein sequences for the presence of Sec amino acid, none of the sequences were found to contain Sec amino acid in it (Supplementary Fig. 4). A protein should not be referred to as a selenoprotein if it does not contain any Sec amino acid in it. Therefore, the protein sequences annotated as “selenocysteine” cannot be considered as a selenoprotein. These contradictions may be the result of an annotation error. Similarly, we also did not find the presence of the proteinogenic amino acid, Pyl encoded by a UAG codon, in any of the fungal proteins analysed so far74. Development of high throughput annotation pipeline can enable us to find the presence of Sec and Pyl amino acids in the fungal proteome in more details.
Fungi encode fewer proteins than plants and animals
Our analysis revealed that fungal proteomes contain an average of 10,345.83 protein sequences per species. Fungal species were previously reported to contain an average of 9113 protein sequences per species75. The previous study, however, was limited to only 143 species which may be the reason why a lower number of protein sequences per species was reported. Plants encode an average of 40,469.47 proteins per species and animals 25,189 per species42. The average of 40,469.83 protein sequences in plants is 391.166% higher than fungi and the average of 25,189 protein sequences per species in animals is 243.47% higher than fungi. This indicates that both plants and animals encode a higher number of protein sequences in their proteome than fungi. No fungal species was found to encode ≥ 40,000 protein sequences in their proteome. The average number of amino acids per fungal protein sequence was calculated to be 459.319. The average number of amino acids per protein sequence in the Ascomycota, Basidiomycota, Chytridiomycota, Microsporidia, Mucoromycota, and Zoopagomycota was 475.406, 446.155, 440.876, 321.063, 413.661, and 407.517, respectively. The average number of amino acids per protein in the Ascomycota species was the highest while the lowest average number was found in Microsporidia (Fig. 6). The average length of plant proteins was reported to be 424.34 amino acids per protein while the average length of all eukaryotic proteins was reported to be 472 amino acids per protein42,75,76. The presence of 459.319 amino acids per protein in fungi falls between the average protein size reported for plants and for all eukaryotic organisms. Longer proteins contain a greater number of conserved domains and display more biological functions. The presence of a lower average number of protein sequences in fungi compared to plants and animals may be the reason that fungi contain a higher number of amino acids per protein. Although the average size of proteins in plants is smaller than in fungi, the protein size is higher in unicellular plant species, such as Chlamydomonas eustigma (576.56), Volvox carteri (568.22), Klebsmordium nitens (538.73), Durio zibethinus (504.36), and Bathycoccus prasinus (521.05)42. This indicates that the average protein size in unicellular organisms is larger than in multicellular organisms (plant, fungi, and animals) and suggests that protein evolution is associated with a reduction in average protein size. Although it is difficult to delineate the exact reasons for the reduction in protein size, evolutionary factors contributing to the reduction include the deletion of exons and/or the fusion of multiple protein domains, both of which may have played a role in determining the size of fungal proteins. In addition, we also did not find any correlation between protein size and pI. Larger proteins are not proportionately associated with a higher pI. Previous studies have reported that the reduction in protein size in the plant genome was partially due to endosymbiosis42,75,77,78. Based on this hypothesis, it seems plausible that the heterotrophic (parasitic and saprophytic) and symbiotic life of fungi may have played an important role in the reduction in protein size that has occurred in the fungi, relative to unicellular plants. The physical structure of a protein is greatly influenced by its primary structure, i.e. the number, composition, and order of the amino acids it contains. Although the chemical environment surrounding a protein plays an important role, the primary sequence of a protein is essential in determining its function. Protein size is an important biochemical parameter as longer proteins can accommodate a greater number of functional protein domains and can therefore display more biological functionality. Shorter proteins/polypeptides possess a limited number of functionalities, while larger proteins, with more functional domains, can serve multiple regulatory functions.
Figure 6.
Average number of amino acids per protein in the fungi. The protein sequences of species within the Ascomycota possess a higher number of amino acids than was found in the species of other phyla. Protein sequences of species within the Microsporidia possess a lower number of amino acids.
Conclusions and future perspectives
A virtual 2-D map of fungal proteomes was constructed and used to evaluate the range of pI and molecular masses present in the proteome of the fungal kingdom. The analysis of the fungal proteome indicated that it does not encode pyrrolysine and selenocysteine amino acids. The average molecular mass of fungal proteins was greater than the average molecular mass of plant proteins, suggesting fungal proteomes are evolutionarily older than plant proteomes. The fungal proteome was dominated by acidic pI proteins, however, the higher percentage of the basic pI proteins in Nosema ceranae may be important and warrants further evaluation to understand its molecular aspects of high pI and possible potential application. Basic pI proteins provide extra positive charges and can serve as an excellent system to study how cells function and operate using a greater number of basic pI proteins. The amino acid composition of the fungal protein will allow us to understand the synonymous codon usage in the protein coding gene and its subsequent evolution. The study can act as a valuable tool for peptide mass finger printing (PMF) and allow to understand the theoretical mass of the peptides. Using this principle, we can understand the molecular mass and pI of sub-proteome and secretome as well. Filamentous fungi secrete a vast number of secreted proteins to carry out their saprophytic lifestyle. Molecular mass and pI of secreted proteins can be valuable to identify the secreted proteins in other organisms. The amino acid composition of fungal proteomes was deciphered using straightforward protein sequences which is useful for finding the homology proteins of known characteristics from an unknown species. However, the sequence similarity model sometimes might fail to work when the query protein did not have significant sequence similarity with the known protein. In this case, the molecular weight and or pI can be very helpful to understand possible function of the protein including its sub-cellular localization.
At the present, we are in the post-genomic era and we have avalanche of protein sequences. We must able to use the newly found novel protein sequences for basic research and biotechnological applications including drug development in large-scale manner. This protein universe of cells can be merged with the systems biology so that it can be incorporated into the gene regulatory network of integrated protein switches. A successful story can be achieved on modular interactions between protein domains and peptide motifs through the integrated approach of molecular weight and pI of proteins. It will be interesting to understand the engineering of single amino acid residue rather than engineering proteins in the cell to understand the role of hyper or hypo amino acid abundance of particular amino acid and its synonymous codon usage and tRNA supply. This will reveal a great deal of evolutionary consequences of synonymous codon usage, tRNA supply, and abundance of particular amino acid.
Material and methods
Sequence retrieval and calculation of molecular weight and pI
The annotated proteome files of the 689 species of fungi deposited in the National Center for Biotechnology Information (NCBI) were downloaded and used for the analyses. The number of amino acids (amino acid composition) in each of the proteome files was determined using Linux-based commands. Molecular masses and isoelectric points of all the proteins in the annotated files were individually analyzed using the IPC in the python module of “protein isoelectric point calculator”79. The generated files for the isoelectric point and molecular mass of the individual species was subsequently converted to an excel file format. All of the quantitative analyses (highest and lowest molecular mass and isoelectric point, composition and abundance of amino acids) were conducted in Microsoft Excel 2016. The virtual 2-D proteome map was constructed using the python-based platform by considering the isoelectric point and molecular weight of 7.15 million protein sequences (x-axis: count 7.209672e + 06 7.209672e + 06, mean: 6.672264e + 00 5.013158e + 01, std: 1.693270e + 00 4.082312e + 01, min: 1.850000e + 00 2.940000e − 03, 25%: 5.347000e + 00 2.371829e + 01, 50%: 6.160000e + 00 4.072890e + 01, 75%: 7.995000e + 00 6.307570e + 01, and max:1.375900e + 01 2.546167e + 03).
Statistical analysis
The principal component analyses (PCA) of the plant proteome were carried out using the statistical software, Unscrambler version 9.7, using the required data from the Excel files. To determine different details on the acidic, basic, and neutral pI of proteins, the data were grouped into Ascomycota, Basidiomycota, Blastocladiomycota, Chytridiomycota, Glomeromycota, Microsporidia, Mucoromycota, Neocallimastigomycota, Ophisthokonta, and Zoopagomycota. The regression analysis, probability plot for the amino acid number, normal probability distribution for acidic and basic pI proteins (%), and Pearson correlation for amino acids were conducted using Past3 software version 1.0.0. Box and whisker plots were constructed using Microsoft Excel 2016.
Supplementary information
Acknowledgements
Authors would like to extend their sincere thanks to Natural and Medical Sciences Research Center, University of Nizwa, Oman for providing require facility to conduct the research. The authors would also like to extend their sincere appreciation to the Deanship of Scientific Research, King Saud University for funding the research group No (RGP-271).
Author contributions
T.K.M.: conceived the idea, collected and annotated the genome sequences, analysed and interpreted the data, drafted and revised the manuscript, A.K.M.: analysed the data; A.K.: analysed the data, A.H. and E.F.A.: revised the manuscript, A.H.: revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tapan Kumar Mohanta and Awdhesh Kumar Mishra.
Contributor Information
Tapan Kumar Mohanta, Email: nostoc.tapan@gmail.com.
Ahmed Al-Harrasi, Email: aharrasi@unizwa.edu.om.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86201-6.
References
- 1.Tang Y, Huang A, Gu Y. Global profiling of plant nuclear membrane proteome in Arabidopsis. Nat. Plants. 2020;6:838–847. doi: 10.1038/s41477-020-0700-9. [DOI] [PubMed] [Google Scholar]
- 2.Joyard J, et al. Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res. 2010;49:128–158. doi: 10.1016/j.plipres.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Kota U, Goshe M. Advances in qualitative and quantitative plant membrane proteomics. Phytochemistry. 2011;72:1040–1060. doi: 10.1016/j.phytochem.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 4.Thelen JJ, Peck SC. Quantitative proteomics in plants: Choices in abundance. Plant Cell. 2007;19:3339–3346. doi: 10.1105/tpc.107.053991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent D, Zivy M. Plant proteome responses to abiotic stress. In: Šamaj J, Thelen JJ, editors. Plant Proteomics. Springer; 2007. pp. 346–364. [Google Scholar]
- 6.Sharma JK, Sihmar M, Santal AR, Singh NP. Impact assessment of major abiotic stresses on the proteome profiling of some important crop plants: A current update. Biotechnol. Genet. Eng. Rev. 2019;35:126–160. doi: 10.1080/02648725.2019.1657682. [DOI] [PubMed] [Google Scholar]
- 7.Mohanta TK, Bae H. Analyses of genomic tRNA reveal presence of novel tRNAs in Oryza sativa. Front. Genet. 2017;8:90. doi: 10.3389/fgene.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schueren F, Thoms S. Functional translational readthrough: A systems biology perspective. PLoS Genet. 2016;12:e1006196–e1006196. doi: 10.1371/journal.pgen.1006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamoto T. Mechanisms of the initiation of protein synthesis: in reading frame binding of ribosomes to mRNA. Mol. Biol. Rep. 2011;38:847–855. doi: 10.1007/s11033-010-0176-1. [DOI] [PubMed] [Google Scholar]
- 10.Kosová K, Vítámvás P, Prasil I, Renaut J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011;74:1301–1322. doi: 10.1016/j.jprot.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Wang X. Protein and proteome atlas for plants under stresses: New highlights and ways for integrated omics in post-genomics era. Int. J. Mol. Sci. 2019;20:5222. doi: 10.3390/ijms20205222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friso G, van Wijk KJ. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015;169:1469–1487. doi: 10.1104/pp.15.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon SJ, Choi EY, Choi YJ, Ahn JH, Park OK. Proteomics studies of post-translational modifications in plants. J. Exp. Bot. 2006;57:1547–1551. doi: 10.1093/jxb/erj137. [DOI] [PubMed] [Google Scholar]
- 14.Yin J, Yi H, Chen X, Wang J. Post-translational modifications of proteins have versatile roles in regulating plant immune responses. Int. J. Mol. Sci. 2019;20:2807. doi: 10.3390/ijms20112807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hvidsten TR, et al. A comprehensive analysis of the structure–function relationship in proteins based on local structure similarity. PLoS ONE. 2009;4:e6266. doi: 10.1371/journal.pone.0006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis R, Moraes I. Structural biology and structure–function relationships of membrane proteins. Biochem. Soc. Trans. 2018;47:47–61. doi: 10.1042/BST20180269. [DOI] [PubMed] [Google Scholar]
- 17.Tramontano A, Cozzetto D. The relationship between protein sequence, structure and function. In: Pifat-Mrzljak G, editor. Supramolecular Structure and Function. Springer; 2004. pp. 15–29. [Google Scholar]
- 18.Chen N, Teng X-L, Xiao X-G. Subcellular localization of a plant catalase-phenol oxidase, AcCATPO, from AMARANTHUS and identification of a non-canonical peroxisome targeting signal. Front. Plant Sci. 2017;8:1345. doi: 10.3389/fpls.2017.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speth EB, Imboden L, Hauck P, He SY. Subcellular localization and functional analysis of the arabidopsis GTPase RabE. Plant Physiol. 2009;149:1824–1837. doi: 10.1104/pp.108.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Ehrhardt DW, Rhee SY. Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol. 2006;141:527–539. doi: 10.1104/pp.106.078881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabsztunowicz M, Koskela MM, Mulo P. Post-translational modifications in regulation of chloroplast function: Recent advances. Front. Plant Sci. 2017;8:240. doi: 10.3389/fpls.2017.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y-C, Peterson SE, Loring JF. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014;24:143–160. doi: 10.1038/cr.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu H, et al. Regulation and role of post-translational modifications of enhancer of zeste homologue 2 in cancer development. Am. J. Cancer Res. 2016;6:2737–2754. [PMC free article] [PubMed] [Google Scholar]
- 24.Schönichen A, Webb BA, Jacobson MP, Barber DL. Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 2013;42:289–314. doi: 10.1146/annurev-biophys-050511-102349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putnam, R. W. Intracellular pH Regulation. in Cell Physiology Source Book (Fourth Edition) (ed. Sperelakis, N. B. T.-C. P. S. B. (Fourth E.) 303–321 (Academic Press, 2012). 10.1016/B978-0-12-387738-3.00017-2
- 26.Yenush L, Merchan S, Holmes J, Serrano R. pH-responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell. Biol. 2005;25:8683–8692. doi: 10.1128/MCB.25.19.8683-8692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odhiambo A, et al. Identification of oxidative post-translational modification of serum albumin in patients with idiopathic pulmonary arterial hypertension and pulmonary hypertension of sickle cell anemia. Rapid Commun. Mass Spectrom. 2007;21:2195–2203. doi: 10.1002/rcm.3074. [DOI] [PubMed] [Google Scholar]
- 28.Höhner R, Aboukila A, Kunz H-H, Venema K. Proton gradients and proton-dependent transport processes in the chloroplast. Front. Plant Sci. 2016;7:218. doi: 10.3389/fpls.2016.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkner G, Horner F, Werdan K, Heldt HW. pH changes in the cytoplasm of the blue-green alga Anacystis nidulans caused by light-dependent proton flux into the thylakoid space. Plant Physiol. 1976;58:717–718. doi: 10.1104/pp.58.6.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savchenko G, Wiese C, Neimanis S, Hedrich R, Heber U. pH regulation in apoplastic and cytoplasmic cell compartments of leaves. Planta. 2000;211:246–255. doi: 10.1007/s004250000280. [DOI] [PubMed] [Google Scholar]
- 31.Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elsliger M-A, Wachter RM, Hanson GT, Kallio K, Remington SJ. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 33.Bagar T, Altenbach K, Read ND, Benčina M. Live-cell imaging and measurement of intracellular ph in filamentous fungi using a genetically encoded ratiometric probe. Eukaryot. Cell. 2009;8:703–712. doi: 10.1128/EC.00333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hesse SJA, Ruijter GJG, Dijkema C, Visser J. Intracellular pH homeostasis in the filamentous fungus Aspergillus niger. Eur. J. Biochem. 2002;269:3485–3494. doi: 10.1046/j.1432-1033.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 35.Martinière A, et al. Uncovering pH at both sides of the root plasma membrane interface using noninvasive imaging. Proc. Natl. Acad. Sci. 2018;115:6488–6493. doi: 10.1073/pnas.1721769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurkdjian A, Guern J. Intracellular pH: Measurement and importance in cell activity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989;40:271–303. doi: 10.1146/annurev.pp.40.060189.001415. [DOI] [Google Scholar]
- 37.Schwartz R, Ting CS, King J. Whole proteome pl values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.GR-1587R. [DOI] [PubMed] [Google Scholar]
- 38.Walter P, Gilmore R, Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984;38:5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- 39.Chung YJ, Krueger C, Metzgar D, Saier MH. Size comparisons among integral membrane transport protein homologues in bacteria, archaea, and eucarya. J. Bacteriol. 2001;183:1012–1021. doi: 10.1128/JB.183.3.1012-1021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberts B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/S0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 41.Le Govic Y, Papon N, Le Gal S, Bouchara J-P, Vandeputte P. Non-ribosomal peptide synthetase gene clusters in the human pathogenic fungus Scedosporium apiospermum. Front. Microbiol. 2019;10:2062. doi: 10.3389/fmicb.2019.02062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohanta TK, Khan AL, Hashem A, Abd-Allah EF, Al-Harrasi A. The molecular mass and isoelectric point of plant proteomes. BMC Genom. 2019;20:631. doi: 10.1186/s12864-019-5983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hupp TR, Sparks A, Lane DP. Edinburgh research explorer small peptides activate the latent sequence-specific DNA binding function of p53 Small peptides activate the latent sequence-specific DNA binding function of ~ 53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 44.Sang Y, Blecha F. Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008;9:227–235. doi: 10.1017/S1466252308001497. [DOI] [PubMed] [Google Scholar]
- 45.Hirakawa Y, Sawa S. Diverse function of plant peptide hormones in local signaling and development. Curr. Opin. Plant Biol. 2019;51:81–87. doi: 10.1016/j.pbi.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Segonzac C, Monaghan J. Modulation of plant innate immune signaling by small peptides. Curr. Opin. Plant Biol. 2019;51:22–28. doi: 10.1016/j.pbi.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell CJ, Stone TA, Deber CM. Peptide-based efflux pump inhibitors of the small multidrug resistance protein from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019;63:e00730–e819. doi: 10.1128/AAC.00730-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilchie, A. L., Hoskin, D. W. & Power Coombs, M. R. Anticancer Activities of Natural and Synthetic Peptides. in Basics for Clinical Application (ed. Matsuzaki, K.) 131–147 (Springer Singapore, 2019). doi:10.1007/978-981-13-3588-4_9 [DOI] [PubMed]
- 49.Hamley IW. Small bioactive peptides for biomaterials design and therapeutics. Chem. Rev. 2017;117:14015–14041. doi: 10.1021/acs.chemrev.7b00522. [DOI] [PubMed] [Google Scholar]
- 50.Lecomte P, Péros J, Blancard D, Bastien N, Délye C. PCR assays that identify the grapevine dieback fungus Eutypa lata. Appl. Environ. Microbiol. 2000;66:4475–4480. doi: 10.1128/AEM.66.10.4475-4480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aufauvre J, et al. Parasite-insecticide interactions: A case study of Nosema ceranae and fipronil synergy on honeybee. Sci. Rep. 2012;2:326. doi: 10.1038/srep00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doublet V, Labarussias M, de Miranda JR, Moritz RFA, Paxton RJ. Bees under stress: Sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ. Microbiol. 2015;17:969–983. doi: 10.1111/1462-2920.12426. [DOI] [PubMed] [Google Scholar]
- 53.Kiraga J, et al. The relationships between the isoelectric point and: Length of proteins, taxonomy and ecology of organisms. BMC Genom. 2007;8:163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knight CG, Kassen R, Hebestreit H, Rainey PB. Global analysis of predicted proteomes: Functional adaptation of physical properties. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8390–8395. doi: 10.1073/pnas.0307270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uversky, V. N. Posttranslational Modification. in (eds. Maloy, S. & Hughes, K. B. T.-B. E. of G. (Second E.) 425–430 (Academic Press, 2013). doi:https://doi.org/10.1016/B978-0-12-374984-0.01203-1
- 56.Müller MM. Post-translational modifications of protein backbones: Unique functions, mechanisms, and challenges. Biochemistry. 2018;57:177–185. doi: 10.1021/acs.biochem.7b00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valnickova Z, et al. Post-translational modifications of human thrombin-activatable fibrinolysis inhibitor (TAFI): Evidence for a large shift in the isoelectric point and reduced solubility upon activation. Biochemistry. 2006;45:1525–1535. doi: 10.1021/bi051956v. [DOI] [PubMed] [Google Scholar]
- 58.Kumar Y, et al. ProteoMod: A new tool to quantitate protein post-translational modifications. Proteomics. 2004;4:1672–1683. doi: 10.1002/pmic.200300778. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka Y, Tanaka R, Kawabata T, Noguchi Y, Himeno M. Lysosomal cysteine protease, cathepsin B, is targeted to lysosomes by the mannose 6-phosphate-independent pathway in rat hepatocytes: Site-specific phosphorylation in oligosaccharides of the proregion1. J. Biochem. 2000;128:39–48. doi: 10.1093/oxfordjournals.jbchem.a022728. [DOI] [PubMed] [Google Scholar]
- 60.Chu A, Matusiewicz N, Stochaj U. Heat-induced nuclear accumulation of hsc70 proteins is regulated by phosphorylation and inhibited in confluent cells. FASEB J. 2001;15:1478–1480. doi: 10.1096/fj.00-0680fje. [DOI] [PubMed] [Google Scholar]
- 61.Gasteiger E, et al. The proteomics protocols handbook. Proteom. Protoc. Handb. 2005;2:571–608. doi: 10.1385/1592598900. [DOI] [Google Scholar]
- 62.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 63.Gonda DK, et al. Universality and stucture of the N-end rule. J. Biol. Chem. 1989;264:16700–16712. doi: 10.1016/S0021-9258(19)84762-2. [DOI] [PubMed] [Google Scholar]
- 64.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 65.Olszowy, P., Burns, A. & Ciborowski, P. Biomolecules. in Proteomic Profiling and Analytical Chemistry (eds. Ciborowski, P. & Silberring, J. B. T.-P. P. and A. C.) 7–24 (Elsevier, 2013). doi:https://doi.org/10.1016/B978-0-444-59378-8.00002-5
- 66.Yu X, Long YC. Crosstalk between cystine and glutathione is critical for the regulation of amino acid signaling pathways and ferroptosis. Sci. Rep. 2016;6:30033. doi: 10.1038/srep30033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gotor C, et al. Low abundance does not mean less importance in cysteine metabolism. Plant Signal. Behav. 2010;5:1028–1030. doi: 10.4161/psb.5.8.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chun SC, Paramasivan M, Chandrasekaran M. Proline accumulation influenced by osmotic stress in arbuscular mycorrhizal symbiotic plants. Front. Microbiol. 2018;9:2525. doi: 10.3389/fmicb.2018.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayat S, et al. Role of proline under changing environments: A review. Plant Signal. Behav. 2012;7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dernovics M, Lobinski R. Characterization of the selenocysteine-containing metabolome in selenium-rich yeast: Part II. On the reliability of the quantitative determination of selenocysteine. J. Anal. Atom. Spectrometry. 2008;23:744–751. doi: 10.1039/b716140a. [DOI] [Google Scholar]
- 71.Mariotti M, Guigó R. Evolution of selenophosphate synthetases: Emergence and relocation of function through independent duplications and recurrent subfunctionalization Running Title : Phylogeny of selenophosphate synthetases Keywords : selenocysteine, gene duplication, sub. Genome Res. 2015;25:1256–1267. doi: 10.1101/gr.190538.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang L, Ni J, Liu Q. Evolution of selenoproteins in the metazoan. BMC Genom. 2012;13:446. doi: 10.1186/1471-2164-13-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mariotti M, Salinas G, Gabaldón T, Gladyshev VN. Utilization of selenocysteine in early-branching fungal phyla. Nat. Microbiol. 2019;4:759–765. doi: 10.1038/s41564-018-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krzycki JA. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 2004;8:484–491. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 75.Ramírez-Sánchez O, Pérez-Rodríguez P, Delaye L, Tiessen A. Plant proteins are smaller because they are encoded by fewer exons than animal proteins. Genom. Proteom. Bioinform. 2016;14:357–370. doi: 10.1016/j.gpb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tiessen A, Pérez-Rodríguez P, Delaye-Arredondo LJ. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res. Notes. 2012;5:85. doi: 10.1186/1756-0500-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sloan DB, Moran NA. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol. Biol. Evol. 2012;29:3781–3792. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozlowski LP. IPC—isoelectric point calculator. Biol. Direct. 2016;11:55. doi: 10.1186/s13062-016-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.