Abstract
Protein cysteine palmitoylation, or S-palmitoylation, has been known for about 40 years and thousands for proteins in humans are known to be modified. Because of the large number of proteins modified, the importance and physiological functions of S-palmitoylation are enormous. However, most of the known physiological functions of S-palmitoylation can be broadly classified into two categories, neurological or immunological. This review provides a summary on the function of S-palmitoylation from the immunological perspective. Several important immune signaling pathways are discussed, including STING, NOD1/2, JAK-STAT in cytokine signaling, T cell receptor signaling, chemotactic GPCR signaling, apoptosis, phagocytosis, and endothelial and epithelial integrity. This review is not meant to be comprehensive, but rather focuses on specific examples to highlight the versatility of palmitoylation in regulating immune signaling, as well as the potential and challenges of targeting palmitoylation to treat immune diseases.
Keywords: Cysteine palmitoylation; immunity; inflammation; ZDHHC; palmitoyltransferase, depalmitoylase; APT1; APT2; ABHD10; ABHD17; STING; NOD1; NOD2; STAT3; IFNAR1; T cell receptor; phagocytosis; CD36; cytokine signaling; G protein-coupled receptor; apoptosis; cell adhesion; cell migration; epithelial barrier; lipid raft; membrane microdomain; phase condensation
Introduction on palmitoylation and regulatory enzymes
S-palmitoylation is the addition of a 16-carbon fatty acyl group to the cysteine side chain of proteins via a labile thioester bond (Figure 1). This modification was initially discovered in the 1980s [1]. Now it has been recognized as a very abundant post-translational modification. The protein palmitoylation database, SwissPalm (https://swisspalm.org/)[2], listed over 3000 human palmitoylated proteins that have been identified via proteomics studies. Several hundreds of them have been biochemically validated.
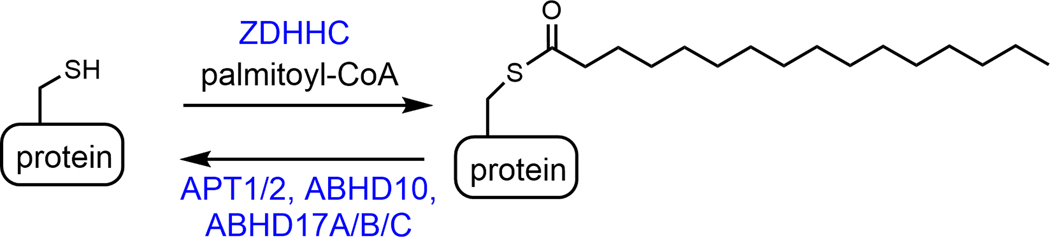
Figure 1.
Reversible protein cysteine palmitoylation catalyzed by palmitoyltransferases (ZDHHCs) and depalmitoylases (APT1/2, ABHD10, and ABHD17).
Palmitoylation is a reversible modification (Figure 1). In mammals, the addition is catalyzed by 23 zinc finger DHHC-type palmitoyltransferases (ZDHHCs) [3]. They are thought to catalyze the transfer of palmitoyl group from palmitoyl-CoA first to the conserve Cys residue in the DHHC motif and then to the cysteine residues in the substrate proteins [4]. They prefer palmitoyl-CoA as substrate, but they can also transfer other structurally similar acyl groups [5]. Thus, in this article, S-palmitoylation is used as a general term to refer to all the similar acylations catalyzed by ZDHHCs. Several depalmitoylases [6, 7], all belongs to the serine hydrolase family, are known to catalyze the removal of S-palmitoylation, including APT1 (LYPLA1) [8], APT2 (LYPLA2) [9, 10], ABHD17A/B/C [11, 12], and ABHD10 [13]. Another protein, PPT1 [14, 15], can also remove S-palmitoylation and mutations in PPT1 causes a neurological disease called, ceroid lipofuscinoses [16]. It is mainly localized inside the lysosomes and is generally believed to remove palmitoyl groups from proteins delivered to the lysosome for degradation [16, 17]. Thus, its regulatory role on protein S-palmitoylation is not well understood. However, a recent study suggest that it regulates Rab7 palmitoylation and thus affects endosome-lysosome fusion [18]. V ATPase subunit V0a1 has also been suggested as PPT1 substrates, but further experimental evidence is needed to confirm this [19].
ZDHHCs are integral membrane proteins with at least four transmembrane helices. They are thought to have distinct membrane localizations, with some mainly in the plasma membrane (ZDHHC5, 20, 23), some mainly in the endoplasmic reticulum or ER (ZDHHC1, 6, 11, 24), some mainly in the Golgi (ZDHHC3, 7, 15, and 18), while most others have several membrane localizations [20, 21]. The membrane localization of ZDHHCs should be taken with flexibility as the localization may be dynamic instead of static and different splicing isoforms or post-translationally modified forms may have different localizations [22, 23]. These localization data are often helpful when trying to determine which ZDHHC is the palmitoyltransferase for a particular substrate protein. In contrast to the ZDHHCs, the known depalmitoylases are all soluble proteins, but some of them, APT1/2, are known to be tethered to membranes via S-palmitoylation [24].
Overview of how palmitoylation affects proteins
While S-palmitoylation is reported to have several different effects on different proteins, including affecting protein-protein interaction, protein stability, and protein aggregation, the most common and direct effect is the increased membrane binding of proteins [21]. This especially true for peripheral membrane proteins, such as Ras subfamily of proteins that are made as soluble proteins, but need to function at the plasma membrane. Such membrane targeting effect of S-palmitoylation sometimes work together with other lipidation events, such as N-terminal glycine myristoylation (e.g. for Fyn and Lyn) or C-terminal prenylation (e.g. for H-Ras and N-Ras).[21] Interestingly, it is reported that a palmitoylation-depalmitoylation cycle maintains specific subcellular distribution patterns of H-Ras and N-Ras [25, 26]. Depalmitoylation redistributes farnesylated Ras in all membranes, followed by palmitoylation and trapping of Ras at the Golgi, from where it is directed to the PM via the secretory pathway. This continuous cycle prevents Ras from nonspecific residence on endomembranes, maintaining the specific intracellular distribution [25, 26].
Obviously, for integral membrane proteins, there is no need to enlist palmitoylation to enhance membrane targeting. However, many integral membrane proteins are known to be palmitoylated [21]. In many cases, the role of palmitoylation is reported to increase the targeting to lipid raft (other names include membrane raft, membrane microdomain, or membrane nanodomain). The nature of lipid raft has been a controversial and evolving topic [27, 28]. I propose that the observation that S-palmitoylation promotes the lipid raft targeting of proteins is likely due to the phase condensation or phase separation of the palmitoylated proteins and their binding partners in the membrane. This is similar to the phase separation phenomena that have been reported for many cytosolic and nuclear proteins [29, 30], but instead this happens in the membrane and promoted by S-palmitoylation. In other words, palmitoylation promotes integral and peripheral membrane proteins to form phase condensation in the membrane. Such phase condensation can concentrate relevant factors in a small space and thus allowing biochemical reactions to occur more efficiently and more specifically. Because of this, forming lipid raft or targeting proteins to lipid raft can be crucial for various cell signaling events that occur at the cell membranes.
Promoting membrane association and formation of lipid rafts may be associated with other effects of S-palmitoylation, such as regulating protein-protein interaction and protein stability. For example, targeting a soluble protein to membranes could increase its interaction with a partner that is membrane associated. Similarly, if two integral membrane proteins are both targeted to the same lipid raft, their interaction can also be increased. If the interacting partner can regulate the ubiquitination or lysosomal targeting of the palmitoylated proteins, then the stability of the protein could also be changed by palmitoylation.
Considering the effect of S-palmitoylation in promoting membrane association and formation of lipid rafts, perhaps it is not surprising that many signaling proteins are regulated by palmitoylation and palmitoylation is well suited for regulating cell signaling events at the cell membranes. Of the ~300 proteins that are biochemically confirmed to be S-palmitoylated, most of the them function in neurological system or immune system (see a list of palmitoylated proteins in a recent review)[21]. Both systems involve numerous cell-cell or cell-environment signaling events that occur at the cell membranes, which may explain why S-palmitoylation is heavily used to regulate neuronal and immune signaling. Here we will focus on the role of palmitoylation in the immune system.
Regulation of STING signaling
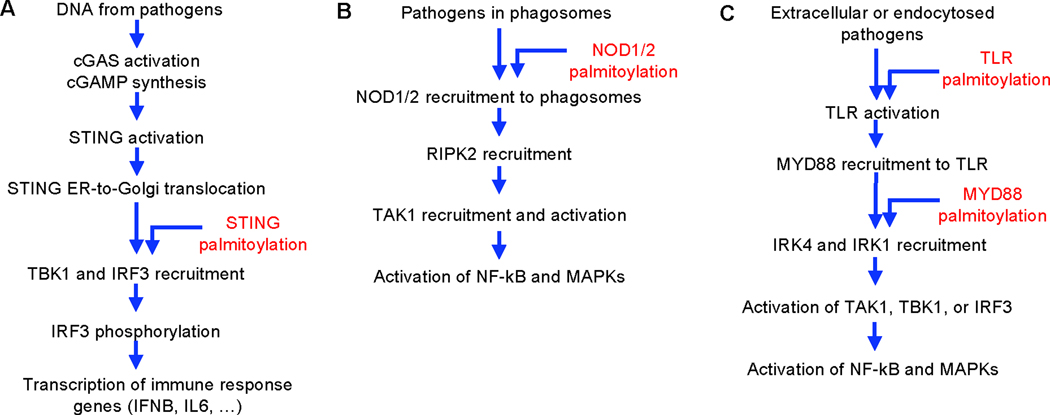
STING is important for type I interferon response against DNA pathogens. It is an integral membrane protein typically associated with the ER. DNA binding to cGAS promotes the synthesis of cyclic dinucleotides, which activates STING and triggers its translocation from the ER to the Golgi, where it activates TBK1, which phosphorylate IRF3 to trigger transcription of immune response genes, including IFNB and IL6 (Figure 2A). STING is later translocated to recycling endosomes and lysosomes. STING palmitoylation occurs at the Golgi mainly on Cys88/91 and is important for activation of TBK1, but not for the trafficking of STING. It is proposed that palmitoylation allows STING to cluster in lipid rafts in the Golgi, allowing it to recruit both TBK1 and IRF3 so that IRF3 phosphorylation could occur (Figure 2A) [31].
Figure 2.
The role of palmitoylation in STING (A), NOD1/2 (B), and TLR (C) signaling pathways. Steps involved in the signaling pathways are listed and the steps that are known to be affected by S-palmitoylation are indicated.
Based on overexpression data, ZDHHC3, 7, and 15 can increase the palmitoylation of STING, suggesting that these ZDHHCs may be responsible for STING palmitoylation [31]. ZDHHC1 and ZDHHC11 are ER associated and have been reported to be important for STING signaling. However, mutants of the DHHC motif can still promote STING signaling, thus, the detailed molecular mechanism is not clear. ZDHHC1 promotes STING dimerization while ZDHHC11 mediates the recruitment of IRF3 to STING [32, 33]. It is not known whether STING can be depalmitoylated by any of the depalmitoylases.
Several small molecules, including C-176 and C-178, are reported to covalent modify Cys91 of mouse STING and thus inhibit STING downstream signaling [34]. These compounds display remarkable selectivity toward STING, considering that there are many proteins containing reactive cysteine residues, and work in mouse models of inflammation. C-176 and C-178 unfortunately do not inhibit human STING, but further refinement of the structures leads to H-151 that can inhibit human STING [34]. Interestingly, 9 or 10-nitro-oleic acid, a nitro fatty acid that can be produced endogenously under inflammation, can also covalently modify STING and inhibit its signaling. This could be a potential feedback regulation to turn down STING signaling [35].
Regulation of NOD1/2 signaling
Nucleotide oligomerization domain (NOD)-like receptors 1 and 2 (NOD1/2) are two related receptors that recognize pathogen-associated molecular patterns (PAMPs), such as peptidoglycan from bacteria [36]. Once activated by ligand, they recruit RIPK2 via their CARD domains. RIPK2 further recruits and activates TAK1, leading to the activation of NF-kB and MAPKs (Figure 2B). NOD1/2 are cytosolic proteins but they are known to be associated with membranes [36]. A recent study demonstrates that NOD1/2 are palmitoylated by ZDHHC5 on multiple cysteine residues (Cys558, 567, 592 on NOD1) and this is important for their membrane recruitment and NF-κB activation [37]. Mutating these palmitoylated cysteine residues to serine disrupts NOD1/2 signaling, but this could be rescued by fusing the first 11 amino acids of the neuronal growth cone protein GAP43 (which is known to be palmitoylated) to NOD1/2. This suggests that the membrane targeting by palmitoylation is important for NOD1/2 signaling. When macrophage cells are infected with Salmonella typhimurium, the bacteria are internalized to phagosomes and NOD1/2 are also recruited to the bacteria-containing phagosomes. Deficiency in NOD1/2 palmitoylation disrupts the recruitment of NOD1/2 to bacteria-containing phagosomes and the activation of NF-kB signaling (Figure 2B). Thus, it appears that palmitoylation is important for targeting NOD1/2 to the bacteria-containing phagosomes, where PAMPs are most concentrated, so that NOD1/2 can efficiently initiate downstream signaling. Several disease-associated mutations in NOD2 are found to be associated with defective S-palmitoylation [37].
Regulation of Toll-like receptor signaling
Like NOD1/2, Toll-like receptors (TLRs) also senses PAMPs to initiate immune responses. However, unlike NOD1/2, TLRs are integral membrane proteins and they sense danger signals that are outside of cellular membranes [38]. Once activated, they recruit adaptor proteins like MYD88, which in turn recruit and activate IRAK4 (Figure 2C). Many of the downstream signaling proteins, such as TAK1, NF-κB, and MAPKs, are shared between the TLR and NOD1/2 signaling pathways. Certain TLRs also signals through TBK1 and IRF3, which overlaps with the STING signaling pathway described above [38].
Because TLRs are integral membrane proteins, unlike NOD1/2, they do not need palmitoylation for membrane targeting. However, in a chemo-proteomic study that identified 563 putative palmitoylation substrates, several TLRs (TLR2, 5 and 10) are identified as potentially palmitoylated proteins [39]. Palmitoylation of TLR2 is mapped to a transmembrane domain-proximal cysteine. Inhibition of TLR2 S-palmitoylation pharmacologically or by cysteine mutagenesis leads to decreased cell surface expression and a decreased inflammatory response to microbial ligands [39]. Thus, palmitoylation can regulate some TLRs.
The adaptor protein for TLR signaling, MYD88 is recently reported to be regulated by S-palmitoylation [40]. ZDHHC6 is responsible for MYD88 palmitoylation on Cys113 and Cys274. Cys113 interaction is important for MYD88 interaction with IRAK4, a serine/threonine kinase important for signaling innate immune responses from TLR (Figure 2C). Interestingly, perturbing endogenous palmitic acid biosynthesis by inhibiting FASN or uptake of exogenous palmitic acid by CD36 decreases MYD88 palmitoylation and TLR signaling [40].
Regulation of phagocytosis
Phagocytosis by immune cells is a way to engulf pathogens or foreign particles and deliver them to lysosomes for destruction. Phagocytosis requires surface receptors that recognize different pathogen or damage-associated patterns. These receptors include immunoglobulin gamma Fc region receptors (FCGRs), complement receptors, and scavenge receptors like CD36 [41]. Particle recognition by these receptors typically leads to the activation of Src family of kinases. Among many downstream signaling pathways, one of them is the activation of Rac GTPases to control actin polymerization to facilitate phagocytosis [41].
One phagocytosis receptor, FCGR2A, is regulated by palmitoylation, which promotes its lipid raft localization [42]. As will be discussed later in T cell signaling, several Src family of kinases are regulated by palmitoylation. Given that some of these kinases are involved in phagocytosis, their palmitoylation could also regulate phagocytosis. An Arf-GAP protein, ASAP2, is reported to be important for FCGR2A mediated phagocytosis. ASAP2 is palmitoylated on Cys86 in a SELK (a selenoprotein) dependent manner. SELK is known to bind ZDHHC6 to promote protein palmitoylation. Thus, ASAP2 is likely palmitoylated by ZDHHC6. Palmitoylation of ASAP2 promotes FCGR-mediated phagocytosis [43].
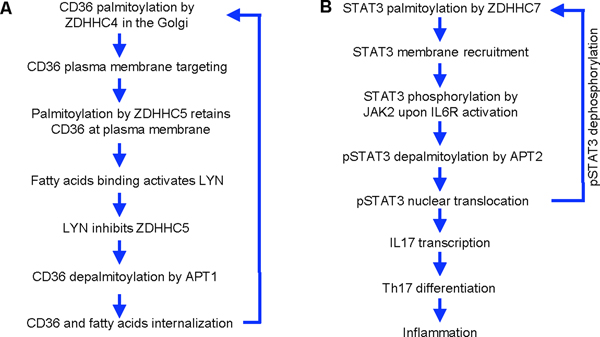
CD36 is a scavenger receptor with important functional implications in immunity, metabolism, and other physiological processes [44]. It can recognize specific oxidized phospholipids, lipoproteins, and free fatty acids, and mediate the phagocytosis of these lipids as well as signaling processes. CD36 is regulated by an interesting and complicated palmitoylation-depalmitoylation cycle (Figure 3A) [45–47]. Four cysteine residues, two at the N-terminal (Cys3 and Cys7) and two at the C-terminal (Cys464 and Cys466) are the palmitoylation sites [45]. CD36 is palmitoylated by DHHC4 in the Golgi, which promotes its export to the plasma membrane [46]. At the plasma membrane, it is kept in the palmitoylated state by DHHC5. However, upon fatty acids binding to CD36, LYN is activated and it phosphorylates DHHC5 on Y91 to inhibit it. This allows ATP1 to depalmitoylate CD36 and promotes its internalization and delivery of fatty acids to lipid droplets. After that, CD36 is repalmitoylated and sent back to the plasma membrane, ready for another cycle [47]. Blocking either palmitoylation or depalmitoylation, or the kinases involved, leads to the inhibition of fatty acid uptake. Whole body knockout of ZDHHC4 or adipose-specific ZDHHC5 knockout both decreased fatty acid uptake and develop hypothermia upon cold exposure [46]. Whether the palmitoylation cycle also affects the signaling/pro-inflammation function of CD36 would be interesting to investigate in the future.
Figure 3.
Palmitoylation-depalmitoylation cycle of CD36 (A) and STAT3 (B). In A, the steps involved in CD36 trafficking are listed to show how the cycle promote the function of CD36 in fatty acid uptake. In B, the steps involved in STAT3 activation and signaling outcomes are listed. The net result of the STAT3 palmitoylation-depalmitoylation cycle is the differentiation of Th17 and inflammation.
CD36 palmitoylation is also regulated by the selenoprotein SELK, which is known to be a partner for ZDHHC6 [48]. CD36 palmitoylation and membrane localization is increased in nonalcoholic steatohepatitis. Inhibition of CD36 palmitoylation not only ameliorates intracellular lipid accumulation, but also inhibits the inflammatory response through the inhibition of the JNK signaling pathway [49]. The fact that CD36 transports fatty acids, produces pro-inflammation signals, is modified by fatty acids, and at the same time provides fatty acids for the modifications of other proteins highlights the complicated connection between lipid metabolism and inflammation.
Regulation of cytokine receptor-mediated signaling
Cytokines, such as interferons and interleukins, plays important roles in immune signaling. Palmitoylation is known to regulate several cytokine-mediated signaling pathways.
IL6 is a cytokine with many important roles in inflammation and immune regulation [50]. It works in many different types of cells, including hepatocytes and CD4 T cells. In CD4 T cells, it is important for the differentiation of Th17 cells. IL6 signals through two proteins, IL6R and Gp130 (gene name IL6ST) and activate STAT3 via JAK2. Recently, it is revealed that several proteins in the IL6 signaling pathway is regulated by palmitoylation. IL6ST is reported to be palmitoylated in axons of dorsal root ganglion neurons. The site of palmitoylation is not identified, but it appears that the knockdown of ZDHHC5 and ZDHHC8 affects IL6ST surface localization and the downstream STAT3 phosphorylation [51].
Interestingly, STAT3 itself is regulated by a palmitoylation-depalmitoylation cycle (Figure 3B) [52]. ZDHHC7 and, to a smaller extent, ZDHHC3, palmitoylate STAT3 on Cys108, promoting its plasma membrane localization, where IL6ST and JAK2 are localized, thus facilitating STAT3 phosphorylation. The phosphorylated STAT3 is then depalmitoylated by APT2, allowing phosphorylated STAT3 to translocate to the nucleus to induce genes that are important for Th17 cell differentiation. Thus, overall, the palmitoylation-depalmitoylation cycle promote STAT3 activation and Th17 cell differentiation. Th17 cells underlies several autoimmune diseases, including inflammatory bowel diseases. Blocking palmitoylation (by knocking out ZDHHC7) or depalmitoylation (by inhibiting APT2) inhibits STAT3 and protected mice in a colitis model [52]. This study highlights the potential of targeting palmitoylation for treating autoimmune diseases.
Type I interferon signaling is also regulated by S-palmitoylation. Type I interferons, including interferon α and β, signals through a heterodimeric receptor that consists of IFNAR1 and IFNAR2. Interferon binding triggers the associated JAK kinases, TYK2 and JAK1, to phosphorylate STAT1 and STAT2. Phosphorylated STAT1 and STAT2 then translocate to the nucleus to promote the transcription of their target genes [53]. Both IFNAR1 and IFNAR2 are palmitoylated [54]. The site of IFNAR1 is identified to be Cys463. This palmitoylation affects STAT1/2 activation, however the exact mechanism is not clear as the localization of IFNAR1 appears to be not affected by Cys463 palmitoylation [54]. Like STAT3, STAT1, which is important for IFNAR1/2 signaling, is also palmitoylated, but the enzymes responsible are not known [52]. Thus, in terms of regulation by S-palmitoylation, interferon signaling has a lot of similarity to IL6 signaling, but much remains to be investigated.
Several interferon-induced proteins, such as interferon-induced transmembrane protein 3 (IFITM3) are also regulated by palmitoylation [55–57] IFITM3 is an endosome- and lysosome-localized protein that restricts numerous virus infections. Its ability to restrict various virus infections requires palmitoylation [58, 59], which is catalyzed redundantly by multiple ZDHHCs, including 3, 7, and 20 [60].
Regulation of chemotactic GPCR signaling
GPCR signaling is important for immune cell chemotaxis, such as migration of monocytes to site of infection and migration of T cells and antigen presenting cells into draining lymph nodes [61]. Chemokines (e.g. CCL5) or specific lipid (e.g. sphingosine 1-phosphate or S1P) signals through GPCR (e.g. CCR5 and S1PR1). Ligand binding to the corresponding GPCR leads to the activation of trimeric Gαβγ proteins. In immune cells, the chemotactic signaling is typically achieved via the Gβγ proteins through the activation of PI3K and PLCβ [61].
S1PR1 is important for immune cell trafficking, such as the normal egress of mature T-cells from the thymus into the blood stream and into peripheral lymphoid organs [62]. ZDHHC5 palmitoylates S1PR1 on three Cys residues at the C-terminal. Palmitoylation is important for the coupling to Gi. Thus, ZDHHC5 can promote immune response via S1PR palmitoylation [63].
CCR5 is a receptor for several chemokines and also serves as a co-receptor of HIV-1. It is palmitoylated on carboxyl-terminal cysteine residues (321, 323, and 324). Inhibition of CCR5 palmitoylation reduces phorbol 12-myristate 13-acetate and CCL5-induced receptor phosphorylation, homologous desensitization, and internalization, but did not impair chemokine-stimulated migration of RBL-2H3 cells [64, 65].
In addition to the GPCRs, several Gα proteins such as Gαq, Gα11, Gαs, and Gαi2 are palmitoylated by ZDHHC3 and ZDHHC7 on one or two N-terminal Cys residues. Palmitoylation is important for the PM localization and PM-Golgi shuttling, as well as GPCR signaling [66]. Thus, S-palmitoylation may also regulate immune cell chemotaxis via the Gα proteins.
Chemotactic GPCR signaling is known to activate Rac small GTPases, which is required for chemotaxis [61]. Rac1 is palmitoylated on Cys178. Non-palmitoylated Rac1 shows decreased GTP loading and lower association with detergent-resistant membranes. Cells lacking RAC1 palmitoylation show spreading and migration defects. These data identify palmitoylation as a mechanism for RAC1 function in actin cytoskeleton remodeling by controlling its membrane partitioning [67]. RAC1 geranylgeranylation and palmitoylation is also reported to be important for RAC1’s ability inhibit RIG1-MAVS signaling, which is important for limiting anti-RNA virus signaling response [68].
RGS (regulator of G protein signaling) proteins can modulate trimeric G protein signaling and are involved in inflammation and immunity [69]. The function of RGS proteins are regulated by palmitoylation. RGS10 Cys60 palmitoylation is important for its activity to inhibit Gsα-mediated signaling [70]. ZDHHC3 and ZDHHC7 palmitoylate RGS4 and stabilizes it [71]. RGS7 family-binding protein (R7BP) regulates the Gi/o signaling by binding to the R7 family (RGS6, RGS7, RGS9, and RGS11) of RGS proteins. R7BP is palmitoylated by ZDHHC2 in neuronal cells. R7BP palmitoylation is dynamic and activation of Gi/o signaling stabilizes palmitoylation while inhibition of Gi/o promotes its depalmitoylation [72]. The palmitoylation of RGS proteins has been mainly investigated in the neuronal system. However, given the involvement of RGS in the immune system, it is very likely that palmitoylation also regulate their function in the immune system.
Regulation of T cell activation
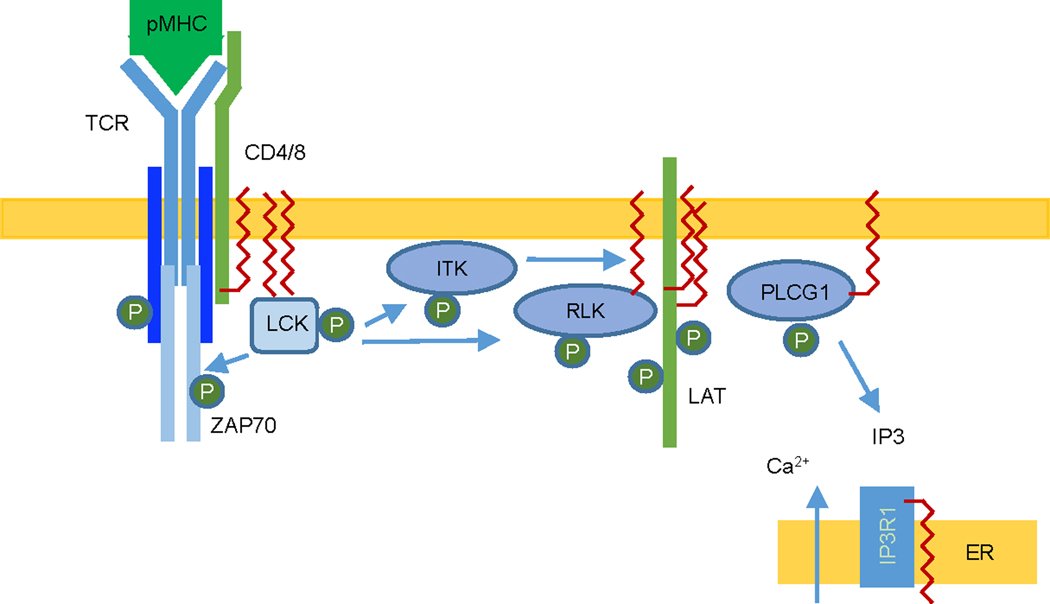
T cells are important for the adaptive immune response. Their development and activation require the T cell receptor (TCR) signaling [73]. In addition to the TCR, TCR signaling also requires CD4 or CD8 co-receptors. Upon binding to MHC-peptide complex on antigen-presenting cells, TCR is activated and clusters to form the immunological synapse. CD4 or CD8 in the immune synapse recruit the Src family tyrosine kinase LCK, leading to the phosphorylation of immunoreceptor tyrosine-based activation motifs (ITAMs) in the cytoplasmic domains of CD3 subunits. This further recruit another tyrosine kinase ZAP70 and the phosphorylation of linker for activation of T cells (LAT) [73]. Downstream from here, many protein kinases can be activated including MAPKs, ITK, VAV1, and PKC, as well as 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 (PLCG1) [73]. Multiple components in the TCR signaling pathway are known to be regulated by S-palmitoylation (Figure 4).
Figure 4.
Multiple proteins in the TCR signaling pathway are regulated by palmitoylation. Key components involved in the signaling pathway are shown. Palmitoylation is indicated by red squiggly lines.
CD4 is palmitoylated on two membrane-proximal Cys residues, Cys396 and Cys399 [74]. The palmitoylation and the association of CD4 with Lck contribute to the enrichment of CD4 in lipid raft. The localization of CD4 in lipid raft also correlates to the ability of CD4 to enhance receptor tyrosine phosphorylation. Palmitoylation thus regulates CD4 signaling [74].
Several Src family tyrosine kinases, including LCK, FYN, and YES, are palmitoylated [75]. LCK is known to be palmitoylated on Cys3 and Cys5, in addition to the N-terminal glycine myristoylation [76]. Palmitoylation is important for LCK plasma membrane targeting and phosphorylation of its substrates, such as CD8 [77–79]. LCK palmitoylation is thought to promote its membrane targeting and also association with lipid raft. It was demonstrated that 13-oxypalmitic acid, which could acylate LCK in place of palmitoylation, allows LCK to target to plasma membrane but not to lipid raft. This inhibits T cell signaling, suggesting the lipid raft association of LCK is important for T cell signaling [80]. Similarly, modification of Fyn with unsaturated or polyunsaturated fatty acids reduces its raft localization and results in decreased T cell signal transduction [81]. Both ZDHHC2 and ZDHHC21 have been reported to palmitoylates LCK [79, 82]. Knockdown of ZDHHC21 leads to decreased T cell activation and decreased T cell marker. Furthermore, ZDHHC21 knockdown also inhibited the differentiation of naïve T cells to Th1 and Th2 [82].
Calcium signaling is important for T cell signaling. The activation of PLCG1 generates inositol triphosphate (IP3), which binds to IP3 receptor (IP3R) to trigger cytosolic calcium increase. PLCG1 is palmitoylated by ZDHHC21 and is dynamically regulated by TCR signaling [82]. The IP3R is also palmitoylated and palmitoylation promote its expression. IP3R palmitoylation is catalyzed by the SELK-dependent ZDHHC6. SELK depletion decreases calcium signaling and impairs immunity [83].
The linker molecule LAT is also regulated by palmitoylation. Palmitoylation on Cys26 and Cys29 is essential for efficient partitioning into membrane microdomains and for its tyrosine phosphorylation [84, 85]. However, there is a report suggesting that the plasma membrane targeting function, but not the lipid raft targeting function, of palmitoylation is important for LAT in TCR signaling [86]. Lack of palmitoylation of LAT is known to lead to anergic T cell [87].
The Tec family kinases are implicated in signaling from lymphocyte antigen receptors and are activated following phosphorylation by Src kinases. For most Tec kinases, this activation requires an interaction between their pleckstrin homology (PH) domains and the products of PI3K. RLK/TXK is a Tec related kinase expressed in T cells that lacks a pleckstrin homology domain, having instead a palmitoylated cysteine-string motif. RLK raft association requires the cysteine-string motif and is independent of PI3K activity [88, 89].
Certain snare proteins like VAMP7, has also be implicated in TCR signaling. VAMP7 is reported to palmitoylated by ZDHHC18 on Cys183. Interestingly this study identifies many proteins that are palmitoylated in a TCR signaling-dependent manner, with VAMP7 being one of them [90]. VAMP7 palmitoylation is important for its trafficking to the Golgi and to localize to the immune synapse upon T cell activation.
CD81, a tetraspanin that is implicated in both B and T cell signaling, is palmitoylated on multiple intracellular cysteines. Interestingly, palmitoylation inhibits its interaction with the epsilon isoform of 14–3-3. Under oxidative stress, palmitoylation is inhibited and the interaction with 14–3-3 increased. CD81 signaling events could be mediated by 14–3-3 adapter proteins, and these signals may be dependent on cellular redox [91].
T cell activation is also controlled by immune checkpoint signaling. Inhibitors of immune check points has been a hot topic for cancer immunotherapy. PD-1 on T cells, upon binding to its ligand PD-L1 and PD-L2, delivers an inhibitory signal to T cells. This contributes to self-tolerance and cancer cell evasion of immune destruction. PD-L1 is reported to be regulated by S-palmitoylation. ZDHHC3 promotes PD-L1 C272 palmitoylation in colorectal cancer cells. Palmitoylation inhibits the mono-ubiquitination of PD-L1 and stabilized it by inhibiting lysosomal degradation. Accordingly, inhibiting PD-L1 palmitoylation increases the killing of these cancer cells by cytotoxic T cells [92]. In breast cancer cells, ZDHHC9 is reported to be the palmitoyltransferase for PD-L1 [93]. PD-1 is also reported to be palmitoylated by ZDHHC9, and palmitoylation is important for its ability to activate mTOR signaling in cancer cells [94].
Regulation of apoptosis signaling
The immune system requires apoptosis because it is important for maintaining self-tolerance and killing infected cells. Many death receptors in the tumor necrosis factor receptor (TNFR) superfamily, such as FAS (other names: CD95/TNFRSF6) and TNFR1 (gene name TNFRSF1A), function at the cell surface and initiate apoptosis by recruiting Fas-associated death domain protein (FADD) and procaspase 8 upon ligand engagement. This leads to the cleavage of procaspase 8 and initiation of the caspase cascade, which is one defining feature of apoptosis [95].
FAS is palmitoylated on Cys199 (Cys194 for mouse FAS). Palmitoylation is important for its lipid raft localization and cell death inducing function [96]. FAS palmitoylation is catalyzed by ZDHHC7 [97]. The same study also revealed that palmitoylation promotes FAS expression by inhibiting its lysosomal degradation. Interestingly, a palmitoylation-defective murine Fas C194V mutant is defective in inducing apoptosis in primary mouse T cells, B cells and dendritic cells, but retains the ability to enhance naive T-cell differentiation, highlighting different effects of palmitoylation on different aspects of FAS function [98].
Other death receptors are also reported to be regulated by S-palmitoylation. DR4 (TNFRSF10A) palmitoylation is required for its raft localization and its ability to transduce death signal induced by TRAIL (TNF-related apoptosis-inducing ligand, TNFSF10) [99]. Death receptor 6 (DR6/TNFRSF21) is also palmitoylated [100]. Recently, TNFR1 is reported to be palmitoylated likely on Cys248 [101]. Mutation of C248 interferes with TNFR1 localization to the plasma membrane. TNFR1 palmitoylation is dynamic and regulated by ligand binding. APT2 is involved in TNFR1 depalmitoylation and TNF-induced signaling [101].
The ligands for death receptors are similarly regulated by palmitoylation. FASL is palmitoylated on Cys82 and palmitoylation promotes FASL targeting to lipid rafts and its death-inducing activity [102]. TNF is palmitoylated on Cys47 and palmitoylation is suggested to regulate TNF lipid raft localization and signaling, but the effect of palmitoylation is not completely understood and requires further investigation [103, 104].
Downstream of FAS signaling may also be regulated by palmitoylation. Caspase 6 is reported to be palmitoylated by ZDHHC17 in neurons [105]. Palmitoylation inhibits caspase 6 activation, which is different from most other cases where palmitoylation promotes the signaling function. In T cells, FAS signaling leads to rapid increase and then decrease in LCK palmitoylation. This dynamic palmitoylation event is regulated by PLCG1. Knockdown of endogenous DHHC21 in Jurkat T cells significantly suppressed both basal and FAS-mediated phosphorylation of LCK, ZAP70, LAT, and PLCG1 [106].
Apoptosis could also be initiated through the intrinsic pathway, which is mainly mediated by the BCL-2-associated X (BAX) protein through permeabilization of the mitochondrial outer membrane and the concomitant release of cytochrome c into the cytosol [95]. BAX is also palmitoylated on Cys126 and palmitoylation promotes its mitochondrial localization and apoptosis induction [107]. Overexpression of several ZDHHCs, including ZDHH3, 7, 11, and 12, can increase BAX palmitoylation and apoptosis. Interestingly, this palmitoylated Cys126 residue (as well as Cys62) can be chemically modified by 2-trans-hexadecenal, which is a byproduct of sphingolipid metabolism. This chemical modification can also potentiate the apoptotic function of BAX [108, 109].
Regulation of endothelial/epithelial barrier integrity, cell adhesion, and cell migration.
The endothelial/epithelial cells form a discrete barrier to components of blood or external environment and thus are important for the immune system. Protein complexes between adjacent cells that create junctions are essential for this barrier function. These junctions control processes such as cell and tissue permeability and cell migration [110]. Many junctional molecules, such as claudin-3 (CLD3), CLD7, CLD14, and junction adhesion molecule 3 (JAM3) are known to be palmitoylated [111–114]. In general, palmitoylation promotes their localization in the tight-junction. The palmitoylation of CLD3 is promoted by ZDHHC12 [113]. JAM3 is palmitoylated by ZDHHC7. Palmitoylation of JAM3 promotes its localization to tight junctions [114]. Desmoglein-2 (DSG2) and plakophilin-3 (PKP3) are two important proteins for forming the desmosomes junctional complex. Both proteins are palmitoylation substrates of ZDHHC5 [23]. ZDHHC5 binds to and palmitoylates GOLGA7B. Palmitoylation of GOLGA7B prevents clathrin-mediated endocytosis of ZDHHC5 and stabilizes it at the plasma membrane. ZDHHC5 and GOLGA7B are required for localization of DSG2 to the plasma membrane and for desmosomal patterning. Loss of ZDHHC5/GOLGA7B causes functional impairments in cell adhesion [23].
Many endothelial proteins also control blood leukocyte transmigration to the site of infection or damage. This typically involves a series of adhesion molecules, such as selectins and integrins, which allow adhesion, crawling, and subsequently migration of cells out of the blood vasculature [115]. Several adhesion proteins are regulated by palmitoylation. Palmitoylation of platelet endothelial cell adhesion molecule 1 (PECAM1 or CD31) promote its lipid raft localization [116]. The palmitoylation of integrin α6β4 by ZDHHC3 is critical for its function, expression, stability, and lipid raft localization [117, 118]. Several tetraspanins (CD9, CD63, CD151), which interact with integrins and affect integrin function, are also palmitoylated [119–121]. ZDHHC2 affects the palmitoylation and stability of tetraspanins CD9 and CD151 [122]. Palmitoylation promoted physical associations between CD9 and CD151, and between α3 integrin, as well as increasing their localization in cell-cell contacts. Palmitoylation also protected CD151 and CD9 from lysosomal degradation [122].
Palmitoylation is also able to regulate signaling events in endothelial cells that contributes to inflammation. ZDHHC21 (and likely also ZDHHC5) can acylate PLCβ and promotes endothelial inflammation [123]. The acylation is increased under systematic inflammation (burn, LPS, or histamine treatment). Acylation promotes PLCβ activity and increases the production of IP3. Accordingly, depletion or inhibition of ZDHHC21 exhibits marked resistance to injury, with reduced plasma leakage, decreased leucocyte adhesion and lung pathology [123]. Similarly, ZDHHC21 deficiency improves gut epithelial barrier dysfunction resulting from burn-induced systemic inflammation [124]. ZDHHC21 palmitoylates eNOS, promoting its membrane localization and NO production [125]. eNOS depalmitoylation is catalyzed by APT1 [126].
Epithelial cell polarity is also important for the integrity of epithelia. One protein that is important for cell polarity is Scribble (SCRIB), a tumor-suppressor protein that plays critical roles in establishing and maintaining epithelial cell polarity. ZDHHC7 acylates while ATP2 deacylates SCRIB. Palmitoylation is important for its correct membrane targeting and cell polarity [127, 128]. SCRIB is also an important regulator of endothelial information and myeloid cell functions including bacterial infection and inflammation [129, 130]. SCRIB interacts directly with the NADPH oxidase (NOX) complex and is required for NOX-induced reactive oxygen species (ROS) generation [130]. Upon bacterial infection, SCRIB localizes to phagosomes and promotes ROS production within phagosomes to kill bacteria. Interestingly, SCRIB loss promotes M1 macrophage polarization and inflammation [130].
Several ZDHHC proteins (ZDHHC13 and ZDHHC21) are reported to have important functions in the skin and hair. Expression of ZDHHC21 in skin is restricted to specific hair lineages. Loss of ZDHHC21 function results in disruption of epidermal homeostasis. The Src-family kinase, FYN, which is involved in keratinocyte differentiation, is a direct palmitoylation target of ZDHHC21 and is mislocalized in mutant follicles [131]. ZDHHC13 is important for maintain skin barrier permeability. Deletion of ZDHHC13 makes mice susceptible to environmental bacterial infection and inflammatory dermatitis [132]. Several palmitoylation substrates have been identified in the skin, including peptidyl arginine deiminase type III, keratin fiber crosslinker transglutaminase 1 [132] and cornifelin [133]. In all these cases, palmitoylation was critical for in vivo protein stability. An interesting substrate of ZDHHC13 is melanocyte-stimulating hormone receptor MC1R [134]. A spontaneous mutation in mouse Zdhhc13 gene increases susceptibility to skin carcinogenesis [135]. In human melanocytes, UV light activates ATR to phosphorylate ZDHHC13 on Ser8, which promotes the interaction between ZDHHC13 and MC1R and the palmitoylation of MC1R [134]. Palmitoylation of MC1R is important for α-melanocyte-stimulating hormone (α-MSH) stimulated cAMP signaling and melanin production, which enhances DNA repair after UV irradiation. Interestingly, MC1R mutants associated with red hair color show reduced palmitoylation and overexpression of ZDHHC13 can rescue their palmitoylation and function. Palmistatin B, a non-specific depalmitoylase inhibitor, is able to increase the palmitoylation of MC1R mutants in red hair color and decrease the incidence of melanoma in a mouse model [134]. It is further demonstrated that APT2 is the depalmitoylation enzyme and inhibition of APT2 with ML349, an APT2 specific inhibitor, decreases incidents of melanoma in a mouse model [136].
Summary and concluding remarks
From the above discussion, it is clear that S-palmitoylation is involved in numerous immune pathways and processes, similar to phosphorylation and ubiquitination. In addition to the wide abundance, there are a few interesting features about palmitoylation. First, even though there are a large number of proteins modified and regulated by palmitoylation, so far there are only 23 palmitoyltransferases known (considering that there are ~500 protein kinases and E3 ubiquitin ligases). Second, the primary known function of palmitoylation can be generalized to either promoting cytosolic proteins’ membrane association or targeting integral or peripheral membrane proteins to specific membrane domains or lipid raft. Other effects of S-palmitoylation, such as regulating protein stability and activity, are likely secondary to the primary effect. Finally, the effect of palmitoylation typically promote the function of the modified proteins and promote the immune signaling response. This makes palmitoylation a promising target for treating immune-related diseases. Promoting palmitoylation may help to fight infections, while inhibiting palmitoylation may help to treat autoimmune disease. Because of this, many translational opportunities will be available to the S-palmitoylation field.
However, to fully take advantage of palmitoylation to treat human diseases, there are also several challenges ahead. For example, given the important role of palmitoylation in various immune pathways as well as other systems such as the brain, to avoid non-desirable side effects, selective inhibition of only one ZDHHC enzyme would be desirable. Currently, there is still no selective inhibitor for ZDHHCs. The most commonly used and potent inhibitor is 2-bromopalmitate, which inhibits most if not all DHHCs and also react with other proteins [21]. The availability of ZDHHC structures will speed up the development of small molecule inhibitors [137]. Another challenge is to understand the complete substrate scope and knockout phenotype of each ZDHHC enzyme so that we can anticipate the functional effect of targeting any particular ZDHHC enzyme. The depalmitoylases provide another opportunity to target palmitoylation. Again, knowing the substrate scope and knockout phenotype would be crucial for future therapeutic development. Currently, there are very good APT1 and APT2 inhibitors and probes available [138–141], which will facilitate future inhibitor development for other depalmitoylases.
Given that palmitoylation typically promotes immune signaling, an interesting question is how the activities of palmitoyltransferases and depalmitoylases are regulated. This is important as too much palmitoylation may lead to uncontrollable inflammation while too little palmitoylation may lead to immune deficiency. Although some regulatory mechanisms are known [142], such as the regulation of ZDHHC13 by phosphorylation [134], the regulation of ZDHHC6 [143] and APT1/APT2 [24] by palmitoylation, much more needs to be learned. Understanding the regulatory mechanisms of palmitoyltransferases and depalmitoylases is another challenge that future research should address.
Addressing these challenges discussed above are within the reach of the S-palmitoylation research community, thanks to the many technologies developed to study S-palmitoylation, such as acyl-biotin exchange [144, 145], acyl-RAC [146], acyl-PEG exchange [57, 147] metabolic labeling with clickable palmitate analogs [148–150], site-specific chemical acylation [151], as well as the ability to quickly generate knockout genes in mice with CRISPR technology [152]. Therefore, we should expect many more exciting new developments in the field in the next decade.
Acknowledgement:
This work is supported in part by fund from HHMI, Cornell University, and NIH (R01CA240529).
Abbreviations
- ABHD
alpha/beta hydrolase domain-containing protein
- Acyl-RAC
S-acyl protein resin-assisted capture
- APT
acyl protein thioesterase
- ARF
ADP-ribosylation factor
- ASAP2
Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 2
- ATR
Ataxia telangiectasia and Rad3-related protein
- BAX
B-cell lymphoma 2-associated X
- CARD
caspase activation and recruitment domains
- CCL5
C-C motif chemokine 5
- CCR5
C-C chemokine receptor type 5
- CLD
claudin
- CRISPR
clustered regularly interspaced short palindromic repeats
- DSG2
Desmoglein-2
- DR4
death receptor 4
- DR6
death receptor 6
- eNOS
Nitric oxide synthase, endothelial
- ER
endoplasmic reticulum
- FADD
Fas-associated death domain protein
- FAS
FS-7 cell-associated surface antigen (gene name TNFRSF6)
- FASL
FAS ligand (gene name FASLG)
- FASN
fatty acid synthase
- FCGR
immunoglobulin gamma Fc region receptor (FCGR)
- GAP
GTPase activating protein
- GAP43
growth-associated protein 43
- GOLGA7B
Golgin subfamily A member 7B
- GPCR
G protein coupled receptor
- HIV-1
human immunodeficiency virus 1
- IL6
Interleukin-6
- IL6R
interleukin-6 receptor subunit alpha
- IL6ST
interleukin-6 receptor subunit beta
- IFITM3
interferon-induced transmembrane protein 3
- IFNAR1/2
Interferon alpha/beta receptor 1/2
- IRAK4
interleukin-1 receptor-associated kinase 4
- IRF3
interferon regulatory factor 3
- ITAM
immunoreceptor tyrosine-based activation motif
- ITK
Interleukin-2-inducible T-cell kinase
- IP3
inositol 1,4,5-triphosphate
- JAK
Janus kinase
- JAM3
junction adhesion molecule 3
- LAT
linker for activation of T cells
- LCK
lymphocyte cell-specific protein-tyrosine kinase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MAVS
Mitochondrial antiviral-signaling protein
- MC1R
melanocyte-stimulating hormone receptor
- mTOR
mammalian target of rapamycin
- MYD88
myeloid differentiation primary response 88
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NOD
nucleotide-binding oligomerization domain-containing protein
- NOX
NADPH oxidase
- PAMPs
pathogen-associated molecular patterns
- PD-1
programmed cell death protein 1 (gene name PDCD1)
- PD-L1/2
programmed cell death 1 ligand ½
- PECAM1
platelet endothelial cell adhesion molecule 1
- PEG
polyethylene glycol
- PI3K
phosphoinositide 3-kinase
- PKC
protein kinase C
- PKP3
plakophilin-3
- PLCβ
phospholipase C beta
- PLCG1
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1
- PM
plasma membrane
- PPT
palmitoyl protein thioesterase
- R7BP
RGS7 family-binding protein
- RGS
regulator of G protein signaling
- RIG1
retinoic acid-inducible gene 1 (gene name DDX58)
- RIPK2
Receptor-interacting serine/threonine-protein kinase 2
- RLK
Resting lymphocyte kinase
- ROS
reactive oxygen species
- S1P
sphingosine 1-phosphate
- S1PR1
Sphingosine 1-phosphate receptor 1
- SCRIB
protein scribble homolog
- SELK
Selenoprotein K
- STAT
Signal transducer and activator of transcription
- STING
Stimulator of interferon genes protein
- TAK1
TGF-beta-activated kinase 1, also called MAP3K7 (Mitogen-activated protein kinase kinase kinase 7)
- TBK1
TANK-binding kinase 1
- TCR
T cell receptor
- TLR
Toll-like receptor
- TNFR
tumor necrosis factor receptor
- TNFRSF
Tumor necrosis factor receptor superfamily member
- TRAIL
TNF-related apoptosis-inducing ligand (gene name TNFSF10)
- TYK2
tyrosine kinase 2
- VAMP7
Vesicle-associated membrane protein 7
- ZAP70
70 kDa zeta-chain associated protein
- ZDHHC
zinc finger DHHC (aspartate-histidine-histidine-cysteine)-type palmitoyltransferases
Footnotes
Conflict of Interest: HL is a founder and consultant for Sedec Therapeutics.
References
- 1.Chen ZQ, Ulsh LS, DuBois G. & Shih TY (1985) Posttranslational processing of p21 ras proteins involves palmitylation of the C-terminal tetrapeptide containing cysteine-186, Journal of virology. 56, 607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J. & van der Goot F. (2015) SwissPalm: Protein Palmitoylation database, F1000Research. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukata M, Fukata Y, Adesnik H, Nicoll RA & Bredt DS (2004) Identification of PSD-95 palmitoylating enzymes, Neuron. 44, 987–96. [DOI] [PubMed] [Google Scholar]
- 4.Jennings BC & Linder ME (2012) DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities, The Journal of biological chemistry. 287, 7236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, Tomkinson NC & Chamberlain LH (2017) Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry, Proceedings of the National Academy of Sciences of the United States of America. 114, E1365-e1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin DT & Conibear E. (2015) Enzymatic protein depalmitoylation by acyl protein thioesterases, Biochemical Society transactions. 43, 193–8. [DOI] [PubMed] [Google Scholar]
- 7.Won SJ, Cheung See Kit M. & Martin BR (2018) Protein depalmitoylases, Critical reviews in biochemistry and molecular biology. 53, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan JA & Gilman AG (1998) A Cytoplasmic Acyl-Protein Thioesterase That Removes Palmitate from G Protein α Subunits and p21RAS, J Biol Chem. 273, 15830–15837. [DOI] [PubMed] [Google Scholar]
- 9.Toyoda T, Sugimoto H. & Yamashita S. (1999) Sequence, expression in Escherichia coli, and characterization of lysophospholipase II, Biochimica et biophysica acta. 1437, 182–93. [DOI] [PubMed] [Google Scholar]
- 10.Tomatis VM, Trenchi A, Gomez GA & Daniotti JL (2010) Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43, PloS one. 5, e15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin DT & Conibear E. (2015) ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization, eLife. 4, e11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K. & Fukata M. (2016) Identification of PSD-95 Depalmitoylating Enzymes, The Journal of neuroscience : the official journal of the Society for Neuroscience. 36, 6431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y, Qiu T, Kathayat RS, Azizi SA, Thorne AK, Ahn D, Fukata Y, Fukata M, Rice PA & Dickinson BC (2019) ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5, Nature chemical biology. 15, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camp LA & Hofmann SL (1993) Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras, The Journal of biological chemistry. 268, 22566–74. [PubMed] [Google Scholar]
- 15.Camp LA, Verkruyse LA, Afendis SJ, Slaughter CA & Hofmann SL (1994) Molecular cloning and expression of palmitoyl-protein thioesterase, The Journal of biological chemistry. 269, 23212–9. [PubMed] [Google Scholar]
- 16.Hellsten E, Vesa J, Olkkonen VM, Jalanko A. & Peltonen L. (1996) Human palmitoyl protein thioesterase: evidence for lysosomal targeting of the enzyme and disturbed cellular routing in infantile neuronal ceroid lipofuscinosis, The EMBO journal. 15, 5240–5. [PMC free article] [PubMed] [Google Scholar]
- 17.Verkruyse LA & Hofmann SL (1996) Lysosomal targeting of palmitoyl-protein thioesterase, The Journal of biological chemistry. 271, 15831–6. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar C, Sadhukhan T, Bagh MB, Appu AP, Chandra G, Mondal A, Saha A. & Mukherjee AB (2020) Cln1-mutations suppress Rab7-RILP interaction and impair autophagy contributing to neuropathology in a mouse model of infantile neuronal ceroid lipofuscinosis, Journal of inherited metabolic disease. 43, 1082–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagh MB, Peng S, Chandra G, Zhang Z, Singh SP, Pattabiraman N, Liu A. & Mukherjee AB (2017) Misrouting of v-ATPase subunit V0a1 dysregulates lysosomal acidification in a neurodegenerative lysosomal storage disease model, Nature communications. 8, 14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohno Y, Kihara A, Sano T. & Igarashi Y. (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins, Biochimica et biophysica acta. 1761, 474–83. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z. & Lin H. (2018) Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies, Chemical reviews. 118, 919–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brigidi GS, Santyr B, Shimell J, Jovellar B. & Bamji SX (2015) Activity-regulated trafficking of the palmitoyl-acyl transferase DHHC5, Nature communications. 6, 8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodley KT & Collins MO (2019) S-acylated Golga7b stabilises DHHC5 at the plasma membrane to regulate cell adhesion, EMBO reports. 20, e47472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB & Mukherjee AB (2013) Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43, The Journal of biological chemistry. 288, 9112–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A. & Bastiaens PI (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms, Science (New York, NY). 307, 1746–52. [DOI] [PubMed] [Google Scholar]
- 26.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H. & Bastiaens PI (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins, Cell. 141, 458–71. [DOI] [PubMed] [Google Scholar]
- 27.Pike LJ (2006) Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function, Journal of lipid research. 47, 1597–8. [DOI] [PubMed] [Google Scholar]
- 28.Lu SM & Fairn GD (2018) Mesoscale organization of domains in the plasma membrane - beyond the lipid raft, Critical reviews in biochemistry and molecular biology. 53, 192–207. [DOI] [PubMed] [Google Scholar]
- 29.Banani SF, Lee HO, Hyman AA & Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry, Nature reviews Molecular cell biology. 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin Y. & Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease, Science (New York, NY). 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- 31.Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, Barber GN, Arai H. & Taguchi T. (2016) Activation of STING requires palmitoylation at the Golgi, Nature communications. 7, 11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Zhou Q, Zhong L, Lin H, Hu MM, Zhou Y, Shu HB & Li S. (2018) ZDHHC11 modulates innate immune response to DNA virus by mediating MITA-IRF3 association, Cellular & molecular immunology. 15, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Lin H, Wang S, Wang S, Ran Y, Liu Y, Ye W, Xiong X, Zhong B, Shu HB & Wang YY (2014) The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling, Cell host & microbe. 16, 450–61. [DOI] [PubMed] [Google Scholar]
- 34.Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, Heymann M, van der Goot FG, Turcatti G, Behrendt R. & Ablasser A. (2018) Targeting STING with covalent small-molecule inhibitors, Nature. 559, 269–273. [DOI] [PubMed] [Google Scholar]
- 35.Hansen AL, Buchan GJ, Rühl M, Mukai K, Salvatore SR, Ogawa E, Andersen SD, Iversen MB, Thielke AL, Gunderstofte C, Motwani M, Møller CT, Jakobsen AS, Fitzgerald KA, Roos J, Lin R, Maier TJ, Goldbach-Mansky R, Miner CA, Qian W, Miner JJ, Rigby RE, Rehwinkel J, Jakobsen MR, Arai H, Taguchi T, Schopfer FJ, Olagnier D. & Holm CK (2018) Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling, Proceedings of the National Academy of Sciences of the United States of America. 115, E7768-e7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caruso R, Warner N, Inohara N. & Núñez G. (2014) NOD1 and NOD2: signaling, host defense, and inflammatory disease, Immunity. 41, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Y, Zheng Y, Coyaud É, Zhang C, Selvabaskaran A, Yu Y, Xu Z, Weng X, Chen JS, Meng Y, Warner N, Cheng X, Liu Y, Yao B, Hu H, Xia Z, Muise AM, Klip A, Brumell JH, Girardin SE, Ying S, Fairn GD, Raught B, Sun Q. & Neculai D. (2019) Palmitoylation of NOD1 and NOD2 is required for bacterial sensing, Science (New York, NY). 366, 460–467. [DOI] [PubMed] [Google Scholar]
- 38.Lim KH & Staudt LM (2013) Toll-like receptor signaling, Cold Spring Harbor perspectives in biology. 5, a011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesarino NM, Hach JC, Chen JL, Zaro BW, Rajaram MV, Turner J, Schlesinger LS, Pratt MR, Hang HC & Yount JS (2014) Chemoproteomics reveals Toll-like receptor fatty acylation, BMC biology. 12, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YC, Lee SE, Kim SK, Jang HD, Hwang I, Jin S, Hong EB, Jang KS & Kim HS (2019) Toll-like receptor mediated inflammation requires FASN-dependent MYD88 palmitoylation, Nature chemical biology. 15, 907–916. [DOI] [PubMed] [Google Scholar]
- 41.Rosales C. & Uribe-Querol E. (2017) Phagocytosis: A Fundamental Process in Immunity, BioMed research international. 2017, 9042851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes NC, Powell MS, Trist HM, Gavin AL, Wines BD & Hogarth PM (2006) Raft localisation of FcgammaRIIa and efficient signaling are dependent on palmitoylation of cysteine 208, Immunology letters. 104, 118–23. [DOI] [PubMed] [Google Scholar]
- 43.Norton RL, Fredericks GJ, Huang Z, Fay JD, Hoffmann FW & Hoffmann PR (2017) Selenoprotein K regulation of palmitoylation and calpain cleavage of ASAP2 is required for efficient FcγR-mediated phagocytosis, Journal of leukocyte biology. 101, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverstein RL & Febbraio M. (2009) CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior, Science signaling. 2, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorne RF, Ralston KJ, de Bock CE, Mhaidat NM, Zhang XD, Boyd AW & Burns GF (2010) Palmitoylation of CD36/FAT regulates the rate of its post-transcriptional processing in the endoplasmic reticulum, Biochimica et biophysica acta. 1803, 1298–307. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Hao JW, Wang X, Guo H, Sun HH, Lai XY, Liu LY, Zhu M, Wang HY, Li YF, Yu LY, Xie C, Wang HR, Mo W, Zhou HM, Chen S, Liang G. & Zhao TJ (2019) DHHC4 and DHHC5 Facilitate Fatty Acid Uptake by Palmitoylating and Targeting CD36 to the Plasma Membrane, Cell reports. 26, 209–221.e5. [DOI] [PubMed] [Google Scholar]
- 47.Hao JW, Wang J, Guo H, Zhao YY, Sun HH, Li YF, Lai XY, Zhao N, Wang X, Xie C, Hong L, Huang X, Wang HR, Li CB, Liang B, Chen S. & Zhao TJ (2020) CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis, Nature communications. 11, 4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meiler S, Baumer Y, Huang Z, Hoffmann FW, Fredericks GJ, Rose AH, Norton RL, Hoffmann PR & Boisvert WA (2013) Selenoprotein K is required for palmitoylation of CD36 in macrophages: implications in foam cell formation and atherogenesis, Journal of leukocyte biology. 93, 771–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao L, Zhang C, Luo X, Wang P, Zhou W, Zhong S, Xie Y, Jiang Y, Yang P, Tang R, Pan Q, Hall AR, Luong TV, Fan J, Varghese Z, Moorhead JF, Pinzani M, Chen Y. & Ruan XZ (2018) CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis, Journal of hepatology. 69, 705–717. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T, Narazaki M. & Kishimoto T. (2014) IL-6 in inflammation, immunity, and disease, Cold Spring Harbor perspectives in biology. 6, a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collura KM, Niu J, Sanders SS, Montersino A, Holland SM & Thomas GM (2020) The palmitoyl acyltransferases ZDHHC5 and ZDHHC8 are uniquely present in DRG axons and control retrograde signaling via the Gp130/JAK/STAT3 pathway, The Journal of biological chemistry. 295, 15427–15437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, Zhou L, Xu Y, Yang M, Xu Y, Komaniecki GP, Kosciuk T, Chen X, Lu X, Zou X, Linder ME & Lin H. (2020) A STAT3 palmitoylation cycle promotes T(H)17 differentiation and colitis, Nature. 586, 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platanias LC (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling, Nature reviews Immunology. 5, 375–86. [DOI] [PubMed] [Google Scholar]
- 54.Claudinon J, Gonnord P, Beslard E, Marchetti M, Mitchell K, Boularan C, Johannes L, Eid P. & Lamaze C. (2009) Palmitoylation of interferon-alpha (IFN-alpha) receptor subunit IFNAR1 is required for the activation of Stat1 and Stat2 by IFN-alpha, The Journal of biological chemistry. 284, 24328–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, López CB & Hang HC (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3, Nature chemical biology. 6, 610–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR & Beard MR (2015) The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry, The Journal of biological chemistry. 290, 25946–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS & Hang HC (2016) Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation, Proceedings of the National Academy of Sciences of the United States of America. 113, 4302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spence JS, He R, Hoffmann HH, Das T, Thinon E, Rice CM, Peng T, Chandran K. & Hang HC (2019) IFITM3 directly engages and shuttles incoming virus particles to lysosomes, Nature chemical biology. 15, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benfield CT, MacKenzie F, Ritzefeld M, Mazzon M, Weston S, Tate EW, Teo BH, Smith SE, Kellam P, Holmes EC & Marsh M. (2020) Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain, Life science alliance. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMichael TM, Zhang L, Chemudupati M, Hach JC, Kenney AD, Hang HC & Yount JS (2017) The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity, The Journal of biological chemistry. 292, 21517–21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamp ME, Liu Y. & Kortholt A. (2016) Function and Regulation of Heterotrimeric G Proteins during Chemotaxis, International journal of molecular sciences. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mendelson K, Evans T. & Hla T. (2014) Sphingosine 1-phosphate signalling, Development (Cambridge, England). 141, 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Badawy SMM, Okada T, Kajimoto T, Ijuin T. & Nakamura SI (2017) DHHC5-mediated palmitoylation of S1P receptor subtype 1 determines G-protein coupling, Scientific reports. 7, 16552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kraft K, Olbrich H, Majoul I, Mack M, Proudfoot A. & Oppermann M. (2001) Characterization of sequence determinants within the carboxyl-terminal domain of chemokine receptor CCR5 that regulate signaling and receptor internalization, The Journal of biological chemistry. 276, 34408–18. [DOI] [PubMed] [Google Scholar]
- 65.Blanpain C, Wittamer V, Vanderwinden JM, Boom A, Renneboog B, Lee B, Le Poul E, El Asmar L, Govaerts C, Vassart G, Doms RW & Parmentier M. (2001) Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways, The Journal of biological chemistry. 276, 23795–804. [DOI] [PubMed] [Google Scholar]
- 66.Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, Perez F. & Fukata M. (2009) Identification of G protein alpha subunit-palmitoylating enzyme, Molecular and cellular biology. 29, 435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Navarro-Lérida I, Sánchez-Perales S, Calvo M, Rentero C, Zheng Y, Enrich C. & Del Pozo MA (2012) A palmitoylation switch mechanism regulates Rac1 function and membrane organization, The EMBO journal. 31, 534–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang S, Harding AT, Sweeney C, Miao D, Swan G, Zhou C, Jiang Z, Fitzgerald KA, Hammer G, Bergo MO, Kroh HK, Lacy DB, Sun C, Glogauer M, Que LG, Heaton NS & Wang D. (2019) Control of antiviral innate immune response by protein geranylgeranylation, Science advances. 5, eaav7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie Z, Chan EC & Druey KM (2016) R4 Regulator of G Protein Signaling (RGS) Proteins in Inflammation and Immunity, The AAPS journal. 18, 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castro-Fernández C, Janovick JA, Brothers SP, Fisher RA, Ji TH & Conn PM (2002) Regulation of RGS3 and RGS10 palmitoylation by GnRH, Endocrinology. 143, 1310–7. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Xie Y, Wolff DW, Abel PW & Tu Y. (2010) DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation, FEBS letters. 584, 4570–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jia L, Linder ME & Blumer KJ (2011) Gi/o signaling and the palmitoyltransferase DHHC2 regulate palmitate cycling and shuttling of RGS7 family-binding protein, The Journal of biological chemistry. 286, 13695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaud G, Lesourne R. & Love PE (2018) Regulatory mechanisms in T cell receptor signalling, Nature reviews Immunology. 18, 485–497. [DOI] [PubMed] [Google Scholar]
- 74.Fragoso R, Ren D, Zhang X, Su MW, Burakoff SJ & Jin YJ (2003) Lipid raft distribution of CD4 depends on its palmitoylation and association with Lck, and evidence for CD4-induced lipid raft aggregation as an additional mechanism to enhance CD3 signaling, Journal of immunology (Baltimore, Md : 1950). 170, 913–21. [DOI] [PubMed] [Google Scholar]
- 75.Koegl M, Zlatkine P, Ley SC, Courtneidge SA & Magee AI (1994) Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif, The Biochemical journal. 303 ( Pt 3), 749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yurchak LK & Sefton BM (1995) Palmitoylation of either Cys-3 or Cys-5 is required for the biological activity of the Lck tyrosine protein kinase, Molecular and cellular biology. 15, 6914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kabouridis PS, Magee AI & Ley SC (1997) S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes, The EMBO journal. 16, 4983–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kosugi A, Hayashi F, Liddicoat DR, Yasuda K, Saitoh S. & Hamaoka T. (2001) A pivotal role of cysteine 3 of Lck tyrosine kinase for localization to glycolipid-enriched microdomains and T cell activation, Immunology letters. 76, 133–8. [DOI] [PubMed] [Google Scholar]
- 79.Zeidman R, Buckland G, Cebecauer M, Eissmann P, Davis DM & Magee AI (2011) DHHC2 is a protein S-acyltransferase for Lck, Molecular membrane biology. 28, 473–86. [DOI] [PubMed] [Google Scholar]
- 80.Hawash IY, Hu XE, Adal A, Cassady JM, Geahlen RL & Harrison ML (2002) The oxygen-substituted palmitic acid analogue, 13-oxypalmitic acid, inhibits Lck localization to lipid rafts and T cell signaling, Biochimica et biophysica acta. 1589, 140–50. [DOI] [PubMed] [Google Scholar]
- 81.Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P. & Resh MD (2001) Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction, The Journal of biological chemistry. 276, 30987–94. [DOI] [PubMed] [Google Scholar]
- 82.Fan Y, Shayahati B, Tewari R, Boehning D. & Akimzhanov AM (2020) Regulation of T cell receptor signaling by protein acyltransferase DHHC21, Molecular biology reports. 47, 6471–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fredericks GJ, Hoffmann FW, Rose AH, Osterheld HJ, Hess FM, Mercier F. & Hoffmann PR (2014) Stable expression and function of the inositol 1,4,5-triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex, Proceedings of the National Academy of Sciences of the United States of America. 111, 16478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang W, Trible RP & Samelson LE (1998) LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation, Immunity. 9, 239–46. [DOI] [PubMed] [Google Scholar]
- 85.Levental I, Lingwood D, Grzybek M, Coskun U. & Simons K. (2010) Palmitoylation regulates raft affinity for the majority of integral raft proteins, Proceedings of the National Academy of Sciences of the United States of America. 107, 22050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hundt M, Harada Y, De Giorgio L, Tanimura N, Zhang W. & Altman A. (2009) Palmitoylation-dependent plasma membrane transport but lipid raft-independent signaling by linker for activation of T cells, Journal of immunology (Baltimore, Md : 1950). 183, 1685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hundt M, Tabata H, Jeon MS, Hayashi K, Tanaka Y, Krishna R, De Giorgio L, Liu YC, Fukata M. & Altman A. (2006) Impaired activation and localization of LAT in anergic T cells as a consequence of a selective palmitoylation defect, Immunity. 24, 513–22. [DOI] [PubMed] [Google Scholar]
- 88.Debnath J, Chamorro M, Czar MJ, Schaeffer EM, Lenardo MJ, Varmus HE & Schwartzberg PL (1999) rlk/TXK encodes two forms of a novel cysteine string tyrosine kinase activated by Src family kinases, Molecular and cellular biology. 19, 1498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chamorro M, Czar MJ, Debnath J, Cheng G, Lenardo MJ, Varmus HE & Schwartzberg PL (2001) Requirements for activation and RAFT localization of the T-lymphocyte kinase Rlk/Txk, BMC immunology. 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morrison E, Wegner T, Zucchetti AE, Álvaro-Benito M, Zheng A, Kliche S, Krause E, Brügger B, Hivroz C. & Freund C. (2020) Dynamic palmitoylation events following T-cell receptor signaling, Communications biology. 3, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clark KL, Oelke A, Johnson ME, Eilert KD, Simpson PC & Todd SC (2004) CD81 associates with 14–3-3 in a redox-regulated palmitoylation-dependent manner, The Journal of biological chemistry. 279, 19401–6. [DOI] [PubMed] [Google Scholar]
- 92.Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, Lu H, Fang C, Zhang Y, Liang L, Zhou X, Wang C, Xue Y, Cui Y. & Xu J. (2019) Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours, Nature biomedical engineering. 3, 306–317. [DOI] [PubMed] [Google Scholar]
- 93.Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu JL, Wei Y, Xia W, Hou J, Qiu Y. & Hung MC (2019) Palmitoylation stabilizes PD-L1 to promote breast tumor growth, Cell research. 29, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao H, Li C, He F, Song T, Brosseau J-P, Wang H, Lu H, Fang C, Shi H, Lan J, Fang J-Y & Xu J. (2021) A peptidic inhibitor for PD-1 palmitoylation targets its expression and functions, RSC Chemical Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu G. & Shi Y. (2007) Apoptosis signaling pathways and lymphocyte homeostasis, Cell research. 17, 759–771. [DOI] [PubMed] [Google Scholar]
- 96.Chakrabandhu K, Hérincs Z, Huault S, Dost B, Peng L, Conchonaud F, Marguet D, He HT & Hueber AO (2007) Palmitoylation is required for efficient Fas cell death signaling, The EMBO journal. 26, 209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossin A, Durivault J, Chakhtoura-Feghali T, Lounnas N, Gagnoux-Palacios L. & Hueber AO (2015) Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability, Cell death and differentiation. 22, 643–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cruz AC, Ramaswamy M, Ouyang C, Klebanoff CA, Sengupta P, Yamamoto TN, Meylan F, Thomas SK, Richoz N, Eil R, Price S, Casellas R, Rao VK, Lippincott-Schwartz J, Restifo NP & Siegel RM (2016) Fas/CD95 prevents autoimmunity independently of lipid raft localization and efficient apoptosis induction, Nature communications. 7, 13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rossin A, Derouet M, Abdel-Sater F. & Hueber AO (2009) Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling, The Biochemical journal. 419, 185–92, 2 p following 192. [DOI] [PubMed] [Google Scholar]
- 100.Klíma M, Zájedová J, Doubravská L. & Andera L. (2009) Functional analysis of the posttranslational modifications of the death receptor 6, Biochimica et biophysica acta. 1793, 1579–87. [DOI] [PubMed] [Google Scholar]
- 101.Zingler P, Särchen V, Glatter T, Caning L, Saggau C, Kathayat RS, Dickinson BC, Adam D, Schneider-Brachert W, Schütze S. & Fritsch J. (2019) Palmitoylation is required for TNF-R1 signaling, Cell communication and signaling : CCS. 17, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guardiola-Serrano F, Rossin A, Cahuzac N, Lückerath K, Melzer I, Mailfert S, Marguet D, Zörnig M. & Hueber AO (2010) Palmitoylation of human FasL modulates its cell death-inducing function, Cell death & disease. 1, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang S, Liu T, Liang H, Zhang H, Yan D, Wang N, Jiang X, Feng W, Wang J, Li P. & Li Z. (2009) Lipid rafts uncouple surface expression of transmembrane TNF-alpha from its cytotoxicity associated with ICAM-1 clustering in Raji cells, Molecular immunology. 46, 1551–60. [DOI] [PubMed] [Google Scholar]
- 104.Poggi M, Kara I, Brunel JM, Landrier JF, Govers R, Bonardo B, Fluhrer R, Haass C, Alessi MC & Peiretti F. (2013) Palmitoylation of TNF alpha is involved in the regulation of TNF receptor 1 signalling, Biochimica et biophysica acta. 1833, 602–12. [DOI] [PubMed] [Google Scholar]
- 105.Skotte NH, Sanders SS, Singaraja RR, Ehrnhoefer DE, Vaid K, Qiu X, Kannan S, Verma C. & Hayden MR (2017) Palmitoylation of caspase-6 by HIP14 regulates its activation, Cell death and differentiation. 24, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akimzhanov AM & Boehning D. (2015) Rapid and transient palmitoylation of the tyrosine kinase Lck mediates Fas signaling, Proceedings of the National Academy of Sciences of the United States of America. 112, 11876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fröhlich M, Dejanovic B, Kashkar H, Schwarz G. & Nussberger S. (2014) S-palmitoylation represents a novel mechanism regulating the mitochondrial targeting of BAX and initiation of apoptosis, Cell death & disease. 5, e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jarugumilli GK, Choi JR, Chan P, Yu M, Sun Y, Chen B, Niu J, DeRan M, Zheng B, Zoeller R, Lin C. & Wu X. (2018) Chemical Probe to Identify the Cellular Targets of the Reactive Lipid Metabolite 2- trans-Hexadecenal, ACS chemical biology. 13, 1130–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen DT, Wales TE, McHenry MW, Engen JR & Walensky LD (2020) Site-Dependent Cysteine Lipidation Potentiates the Activation of Proapoptotic BAX, Cell reports. 30, 3229–3239.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zihni C, Mills C, Matter K. & Balda MS (2016) Tight junctions: from simple barriers to multifunctional molecular gates, Nature reviews Molecular cell biology. 17, 564–80. [DOI] [PubMed] [Google Scholar]
- 111.Heiler S, Mu W, Zöller M. & Thuma F. (2015) The importance of claudin-7 palmitoylation on membrane subdomain localization and metastasis-promoting activities, Cell communication and signaling : CCS. 13, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Van Itallie CM, Gambling TM, Carson JL & Anderson JM (2005) Palmitoylation of claudins is required for efficient tight-junction localization, Journal of cell science. 118, 1427–36. [DOI] [PubMed] [Google Scholar]
- 113.Yuan M, Chen X, Sun Y, Jiang L, Xia Z, Ye K, Jiang H, Yang B, Ying M, Cao J. & He Q. (2020) ZDHHC12-mediated claudin-3 S-palmitoylation determines ovarian cancer progression, Acta pharmaceutica Sinica B. 10, 1426–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aramsangtienchai P, Spiegelman NA, Cao J. & Lin H. (2017) S-Palmitoylation of Junctional Adhesion Molecule C Regulates Its Tight Junction Localization and Cell Migration, The Journal of biological chemistry. 292, 5325–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nourshargh S. & Alon R. (2014) Leukocyte migration into inflamed tissues, Immunity. 41, 694–707. [DOI] [PubMed] [Google Scholar]
- 116.Sardjono CT, Harbour SN, Yip JC, Paddock C, Tridandapani S, Newman PJ & Jackson DE (2006) Palmitoylation at Cys595 is essential for PECAM-1 localisation into membrane microdomains and for efficient PECAM-1-mediated cytoprotection, Thrombosis and haemostasis. 96, 756–66. [PubMed] [Google Scholar]
- 117.Gagnoux-Palacios L, Dans M, van’t Hof W, Mariotti A, Pepe A, Meneguzzi G, Resh MD & Giancotti FG (2003) Compartmentalization of integrin alpha6beta4 signaling in lipid rafts, The Journal of cell biology. 162, 1189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sharma C, Rabinovitz I. & Hemler ME (2012) Palmitoylation by DHHC3 is critical for the function, expression, and stability of integrin α6β4, Cellular and molecular life sciences : CMLS. 69, 2233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA & Hemler ME (2002) Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology, Molecular biology of the cell. 13, 767–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berditchevski F, Odintsova E, Sawada S. & Gilbert E. (2002) Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling, The Journal of biological chemistry. 277, 36991–7000. [DOI] [PubMed] [Google Scholar]
- 121.Israels SJ & McMillan-Ward EM (2010) Palmitoylation supports the association of tetraspanin CD63 with CD9 and integrin alphaIIbbeta3 in activated platelets, Thrombosis research. 125, 152–8. [DOI] [PubMed] [Google Scholar]
- 122.Sharma C, Yang XH & Hemler ME (2008) DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151, Molecular biology of the cell. 19, 3415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beard RS Jr., Yang X, Meegan JE, Overstreet JW, Yang CG, Elliott JA, Reynolds JJ, Cha BJ, Pivetti CD, Mitchell DA, Wu MH, Deschenes RJ & Yuan SY (2016) Palmitoyl acyltransferase DHHC21 mediates endothelial dysfunction in systemic inflammatory response syndrome, Nature communications. 7, 12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Haines RJ, Wang CY, Yang CGY, Eitnier RA, Wang F. & Wu MH (2017) Targeting palmitoyl acyltransferase ZDHHC21 improves gut epithelial barrier dysfunction resulting from burn-induced systemic inflammation, American journal of physiology Gastrointestinal and liver physiology. 313, G549–g557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fernández-Hernando C, Fukata M, Bernatchez PN, Fukata Y, Lin MI, Bredt DS & Sessa WC (2006) Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase, The Journal of cell biology. 174, 369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yeh DC, Duncan JA, Yamashita S. & Michel T. (1999) Depalmitoylation of endothelial nitric-oxide synthase by acyl-protein thioesterase 1 is potentiated by Ca(2+)-calmodulin, The Journal of biological chemistry. 274, 33148–54. [DOI] [PubMed] [Google Scholar]
- 127.Chen B, Zheng B, DeRan M, Jarugumilli GK, Fu J, Brooks YS & Wu X. (2016) ZDHHC7-mediated S-palmitoylation of Scribble regulates cell polarity, Nature chemical biology. 12, 686–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hernandez JL, Davda D, Cheung See Kit M, Majmudar JD, Won SJ, Gang M, Pasupuleti SC, Choi AI, Bartkowiak CM & Martin BR (2017) APT2 Inhibition Restores Scribble Localization and S-Palmitoylation in Snail-Transformed Cells, Cell chemical biology. 24, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kruse C, Kurz AR, Pálfi K, Humbert PO, Sperandio M, Brandes RP, Fork C. & Michaelis UR (2015) Polarity Protein Scrib Facilitates Endothelial Inflammatory Signaling, Arteriosclerosis, thrombosis, and vascular biology. 35, 1954–62. [DOI] [PubMed] [Google Scholar]
- 130.Zheng W, Umitsu M, Jagan I, Tran CW, Ishiyama N, BeGora M, Araki K, Ohashi PS, Ikura M. & Muthuswamy SK (2016) An interaction between Scribble and the NADPH oxidase complex controls M1 macrophage polarization and function, Nature cell biology. 18, 1244–1252. [DOI] [PubMed] [Google Scholar]
- 131.Mill P, Lee AW, Fukata Y, Tsutsumi R, Fukata M, Keighren M, Porter RM, McKie L, Smyth I. & Jackson IJ (2009) Palmitoylation regulates epidermal homeostasis and hair follicle differentiation, PLoS genetics. 5, e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen LY, Yang-Yen HF, Tsai CC, Thio CL, Chuang HL, Yang LT, Shen LF, Song IW, Liu KM, Huang YT, Liu FT, Chang YJ, Chen YT & Yen JJY (2017) Protein Palmitoylation by ZDHHC13 Protects Skin against Microbial-Driven Dermatitis, The Journal of investigative dermatology. 137, 894–904. [DOI] [PubMed] [Google Scholar]
- 133.Liu KM, Chen YJ, Shen LF, Haddad ANS, Song IW, Chen LY, Chen YJ, Wu JY, Yen JJY & Chen YT (2015) Cyclic Alopecia and Abnormal Epidermal Cornification in Zdhhc13-Deficient Mice Reveal the Importance of Palmitoylation in Hair and Skin Differentiation, The Journal of investigative dermatology. 135, 2603–2610. [DOI] [PubMed] [Google Scholar]
- 134.Chen S, Zhu B, Yin C, Liu W, Han C, Chen B, Liu T, Li X, Chen X, Li C, Hu L, Zhou J, Xu ZX, Gao X, Wu X, Goding CR & Cui R. (2017) Palmitoylation-dependent activation of MC1R prevents melanomagenesis, Nature. 549, 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Perez CJ, Mecklenburg L, Jaubert J, Martinez-Santamaria L, Iritani BM, Espejo A, Napoli E, Song G, Del Río M, DiGiovanni J, Giulivi C, Bedford MT, Dent SYR, Wood RD, Kusewitt DF, Guénet JL, Conti CJ & Benavides F. (2015) Increased Susceptibility to Skin Carcinogenesis Associated with a Spontaneous Mouse Mutation in the Palmitoyl Transferase Zdhhc13 Gene, The Journal of investigative dermatology. 135, 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen S, Han C, Miao X, Li X, Yin C, Zou J, Liu M, Li S, Stawski L, Zhu B, Shi Q, Xu ZX, Li C, Goding CR, Zhou J. & Cui R. (2019) Targeting MC1R depalmitoylation to prevent melanomagenesis in redheads, Nature communications. 10, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rana MS, Kumar P, Lee CJ, Verardi R, Rajashankar KR & Banerjee A. (2018) Fatty acyl recognition and transfer by an integral membrane S-acyltransferase, Science (New York, NY). 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adibekian A, Martin BR, Chang JW, Hsu K-L, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Hodder PS, Rosen H. & Cravatt BF (2012) Confirming Target Engagement for Reversible Inhibitors in Vivo by Kinetically Tuned Activity-Based Probes, J Am Chem Soc. 134, 10345–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Won SJ, Davda D, Labby KJ, Hwang SY, Pricer R, Majmudar JD, Armacost KA, Rodriguez LA, Rodriguez CL, Chong FS, Torossian KA, Palakurthi J, Hur ES, Meagher JL, Brooks CL 3rd, Stuckey JA & Martin BR (2016) Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2), ACS chemical biology. 11, 3374–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kathayat RS, Elvira PD & Dickinson BC (2017) A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling, Nature chemical biology. 13, 150–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Azizi SA, Kathayat RS & Dickinson BC (2019) Activity-Based Sensing of S-Depalmitoylases: Chemical Technologies and Biological Discovery, Accounts of chemical research. 52, 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zmuda F. & Chamberlain LH (2020) Regulatory effects of post-translational modifications on zDHHC S-acyltransferases, The Journal of biological chemistry. 295, 14640–14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Abrami L, Dallavilla T, Sandoz PA, Demir M, Kunz B, Savoglidis G, Hatzimanikatis V. & van der Goot FG (2017) Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade, eLife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Drisdel RC & Green WN (2004) Labeling and quantifying sites of protein palmitoylation, BioTechniques. 36, 276–85. [DOI] [PubMed] [Google Scholar]
- 145.Wan J, Roth AF, Bailey AO & Davis NG (2007) Palmitoylated proteins: purification and identification, Nature protocols. 2, 1573–84. [DOI] [PubMed] [Google Scholar]
- 146.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA & Stamler JS (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture, Nature biotechnology. 27, 557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Speca DJ & Diaz E. (2020) Acyl-PEGyl Exchange Gel Shift Assay for Quantitative Determination of Palmitoylation of Brain Membrane Proteins, Journal of visualized experiments : JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ & Ploegh HL (2007) Chemical probes for the rapid detection of Fatty-acylated proteins in Mammalian cells, J Am Chem Soc. 129, 2744–5. [DOI] [PubMed] [Google Scholar]
- 149.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E. & Hang HC (2009) Robust fluorescent detection of protein fatty-acylation with chemical reporters, J Am Chem Soc. 131, 4967–75. [DOI] [PubMed] [Google Scholar]
- 150.Martin BR & Cravatt BF (2009) Large-scale profiling of protein palmitoylation in mammalian cells, Nature methods. 6, 135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li Y, Wang S, Chen Y, Li M, Dong X, Hang HC & Peng T. (2020) Site-specific chemical fatty-acylation for gain-of-function analysis of protein S-palmitoylation in live cells, Chemical communications (Cambridge, England). 56, 13880–13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA & Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems, Science (New York, NY). 339, 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]