Abstract
Early recognition of severe forms of coronavirus disease 2019 (COVID-19) is essential for an opportune and effective intervention, reducing life-risking complications. An altered inflammatory immune response seems to be associated with COVID-19’s pathogenesis and progression to severity. Here we demonstrate the utility of early nasopharyngeal swab samples for detection of the early expression of immune markers and the potential value of CCL2/MCP-1 in predicting disease outcome.
Keywords: COVID-19, chemokines, cytokines, inflammation, prognostic biomarkers
Coronavirus disease 2019 (COVID-19) is an emerging infectious disease that is highly pathogenic and is presently threatening global health. Cuba reported the first confirmed COVID-19 cases on March 11, 2020, and autochthonous transmission on April 7. The first confirmed cases in Cuba were diagnosed and hospitalized at the Institute of Tropical Medicine Pedro Kourí (IPK) at the Cuban National Center of Reference for Infectious Diseases.
Severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) virus, the etiological agent of COVID-19, produces both symptomatic and asymptomatic infections. As a result, patients can be asymptomatic or have mild respiratory disease, acute respiratory distress syndrome (ARDS), or pneumonia of varying degrees of severity [1].
SARS-CoV-2 virus shows, similarly to Middle Eastern respiratory syndrome (MERS) coronavirus infection, a high viral replication in the upper airway epithelial cells, resulting in enhanced production of multiple cytokines and chemokines [2, 3]. Some proinflammatory mediators, like TNFα, CCL2, and CCL3, could be directly responsible for vascular leakage and alveolar edema, ultimately causing hypoxia, ARDS, and potentially death, through the apoptosis of both lung epithelial and endothelial cells [4].
In view of this, we considered the possible utility of early nasopharyngeal swab samples to detect the presence of messenger RNA of proinflammatory and regulatory mediators already associated with COVID-19 disease, like TNFα, CCL2/MCP-1, CCL3/MIP-1α, and TGFβ and IL-10, respectively. We hypothesized that the activity of this set of cytokines/chemokines could trigger/suppress severe outcomes of infection, resulting in confirmed COVID-19 patients with asymptomatic infections or different clinical evolutions.
We found significant differences between the groups with favorable and unfavorable clinical evolutions, suggesting the value of early detection of TNFα, CCL2/MCP-1, and CCL3/MIP-1α mRNA in nasopharyngeal swab samples and the predictive value of CCL2/MCP-1 for the COVID-19 clinical outcome.
METHODS
Clinical Samples
Nasopharyngeal samples were collected separately with sterile polyester-tipped swabs (Puritan Medical Products Co., LLC, Guilford, ME, USA) following the Pan American Health Organization/World Health Organization (WHO) Laboratory Guidelines for Detection and Diagnosis of the Novel Coronavirus (2019‐nCoV). Both swabs of each patient (nasophayngeal samples) were placed into the same tube with universal transport medium in the collection tube (Puritan UniTranz-RT Transport System).
Sample manipulation was performed in a biosafety class II lab. Total RNA was isolated from nasopharyngeal swabs using the RNeasy Mini QIAcube Kit in the QIAcube instrument (Qiagen, Hilden, Germany), following the supplier’s instructions.
COVID-19 infection was confirmed by real-time reverse transcription polymerase chain reaction (RT-PCR) assay for nasopharyngeal swab specimens, following WHO guidelines for qRT-PCR [5].
Isolated RNA from nasopharyngeal swab samples from 54 cases received at the Reference Laboratories of the Pedro Kourí Institute of Tropical Medicine as part of COVID-19 surveillance in the period of March 11 to April 7 was studied. Thirty-eight samples were confirmed as COVID-19-positive. Patients were admitted at the Medical Attention Branch of the IPK. Of these patients, 12 remained asymptomatic and 26 developed COVID-19 symptoms (fever, cough, myalgia, diarrhea rhinorrhea, sore throat, vomiting, asthenia, dyspnea, and respiratory distress) (Table 1).
Table 1.
Characterization of the Studied Subjects
| Clinical Classification | Healthy (n = 16) | Asymptomatic (n = 12) | Mild Disease (n = 11) | Very Symptomatic (n = 9) | Severe (n = 3) | Fatal (n = 3) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Groups | Controls (n = 16) | Favorable Evolution (n = 23) | Unfavorable Evolution (n = 15) | ||||||||
| Age, y | No. | % | No. | % | No. | % | |||||
| 15–54 | 9 | 56.3 | 7 | 58.3 | 8 | 61.5 | |||||
| 55–64 | 5 | 31.2 | 1 | 8.3 | 3 | 23.1 | |||||
| >65 | 2 | 12.5 | 4 | 33.3 | 2 | 15.4 | |||||
| Sex: female | 8 | 50 | 6 | 50 | 5 | 38.5 | |||||
| Sex: male | 8 | 50 | 6 | 50 | 8 | 61.5 | |||||
| Comorbidities | Healthy | Asymptomatic | Mild disease | Very symptomatic | Severe | Fatal | |||||
| Hypertension | - | 1 | 3 | 2 | 2 | 1 | |||||
| Cardiopathy | - | 0 | 0 | 1 | 1 | 1 | |||||
| Diabettes mellitus | - | 0 | 0 | 0 | 1 | 2 | |||||
| Bronchial asthma | - | 2 | 1 | 1 | 1 | 1 | |||||
| Obesity | - | 0 | 0 | 0 | 0 | 1 | |||||
| Other (autoimmunity, epilepsy, cancer, HIV) | - | 5 | 0 | 0 | 0 | 1 | |||||
| Signs/symptoms | Healthy | Asymptomatic | Mild disease | Very symptomatic | Severe | Fatal | |||||
| No. | % | No. | % | No. | % | No. | % | ||||
| Fever | 0 | 0 | 7 | 53.8 | 9 | 100 | 3 | 100 | 3 | 60 | |
| Cough | 0 | 0 | 8 | 61.5 | 9 | 100 | 3 | 100 | 4 | 80 | |
| Sore throat | 0 | 0 | 3 | 23.1 | 2 | 22.2 | 3 | 100 | 1 | 20 | |
| Rhinorrhea | 0 | 0 | 3 | 23.1 | 1 | 11.1 | 0 | 0 | 0 | 0 | |
| Myalgia | 0 | 0 | 0 | 0 | 4 | 44.4 | 1 | 33.3 | 1 | 20 | |
| Asthenia | 0 | 0 | 0 | 0 | 5 | 55.5 | 1 | 33.3 | 4 | 80 | |
| Dyspnea | 0 | 0 | 0 | 0 | 4 | 44.4 | 1 | 33.3 | 4 | 80 | |
| Diarrhea | 0 | 0 | 1 | 7.6 | 3 | 33.3 | 0 | 0 | 1 | 20 | |
| Vomiting | 0 | 0 | 0 | 0 | 1 | 11.1 | 0 | 0 | 0 | 0 | |
| Respiratory distressa | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 100 | 5 | 100 | |
Abbreviations: COVID-19, coronavirus disease 2019; ICU, intensive care unit; PaO2/FiO2, ratio of arterial oxygen partial pressure to fraction of inspired oxygen.
aRespiratory distress: Respiratory frequency > 30/min, O2 saturation index <93%, PaO2/FiO2 ratio <300.
Symptomatic cases were classified according to the number of symptoms and the presence of symptoms previously associated with severity in Cuban patients (respiratory distress, dyspnea, and asthenia). Patients with 1 or 2 symptoms, excluding those associated with severity, were classified as having mild disease (11 patients). Patients with 5 or more symptoms, including asthenia and dyspnea, were classified as very symptomatic (9 patients). Patients with respiratory distress (respiratory frequency >30/min, O2 saturation index <93%, PaO2/FiO2 Ratio <300) and intensive care admission but who progressed to the recovery phase were classified as severe (3 patients); those with respiratory distress and intensive care admission who died were classified as fatal (3 patients). None of the asymptomatic or mild disease cases showed levels of C-reactive protein >20 mg/L, levels of ferritin >400 μg/L, or lymphopenia (lymphocyte count <1.0 ×109/L). To identify biomarkers that discriminate between a favorable and unfavorable clinical evolution, we joined asymptomatic subjects and patients with mild disease in the category of “favorable evolution” and all those patients who developed a more severe picture (very symptomatic, complicated, and fatal cases) in the category “unfavorable evolution” (Table 1).
All swab samples from symptomatic patients were collected between the first and fifth day after onset of COVID-19 symptoms. Samples from 16 healthy individuals confirmed as negative for SARS-CoV-2 were used as controls. The information about age, sex, days from symptom onset to sample collection, and comorbidities of the studied subjects are shown in Table 1.
Cytokine Gene Expression Analysis
cDNA was synthesized from mRNA with poly(dT) primers and Superscript II reverse transcriptase assay (Life Technologies, Rockville, MD, USA). Quantitative real-time PCR (qPCR) was performed with Rotor-Gene Q 5plex HRM (Qiagen) in 36-well Gene Discs using the QuantiFast SYBR Green RT-PCR Kit (Qiagen) following the manufacturer’s instructions. The PCR protocol is described in detail in the Supplementary Data.
Gene expression variations of TNFα, CCL2/MCP-1, CCL3/MIP-1 α, TGFβ, and IL10 were evaluated in terms of fold induction with respect to the housekeeping gene hypoxanthine phosphoribosyltransferase-1 (HPRT-1) by the 2-∆∆CT method. Experiments were conducted in triplicate. The sequence of primers used in this work is described in the Supplementary Data.
Statistical Analysis
The distribution of the data was tested using the Kolmogorov-Smirnov test (data not normally distributed). The Kruskal-Wallis nonparametric test was used to determine if there were significant differences in the gene expression of each cytokine among the 3 groups of cases (controls, cases with favorable evolution, and cases with unfavorable evolution). If there was statistical significance, we proceed to Dunn’s correction for multiple comparisons. For analyzing the differences in the gene expression of each cytokine according to the time from symptom onset to sample collection and the association of cytokine gene expression with symptoms, we used the nonparametric Wilcoxon-Mann-Whitney U mean rank test. Graphics, shown as individual dots and with data on a logarithmic scale, were made with the program GraphPad Prism 5 for Windows, version 5.01. Data are displayed as median and range. Correlations between variables were evaluated with the Spearman correlation test. The values of the cytokines were log-transformed to address skewed distribution before univariate and multivariate logistic regression analysis. Data analyses were considered statistically significant when P < .05. Calculations were made with the Statistical Package for Social Sciences, version 11.5, and R, version 4.0.1, for Microsoft Windows 10.
Patient Consent Statement
Written informed consent was obtained from each individual upon enrollment in the study. This study was conducted according to the Declaration of Helsinki. The design of the work was approved by the Institutional Ethical Review Committee of the Institute of Tropical Medicine “Pedro Kourí” and the Cuban Ministry of Public Health.
RESULTS
The objective of this study was to explore the early expression of TNFα, CCL2/MCP-1, CCL3/MIP-1α, IL-10, and TGFβ, which were previously associated with the uncontrolled cytokine response in ARDS pathogenesis produced by human pathogenic coronaviruses, taking advantage of the nasopharyngeal swab sample, which is mandatory for the diagnosis of COVID-19 in suspected cases in Cuba.
To determine the possible association of early gene expression of immune mediators in nasopharyngeal swab samples with clinical evolution, we studied patients with a mild or severe COVID-19 clinical picture and asymptomatic subjects. SARS-CoV-2-negative individuals were included as controls.
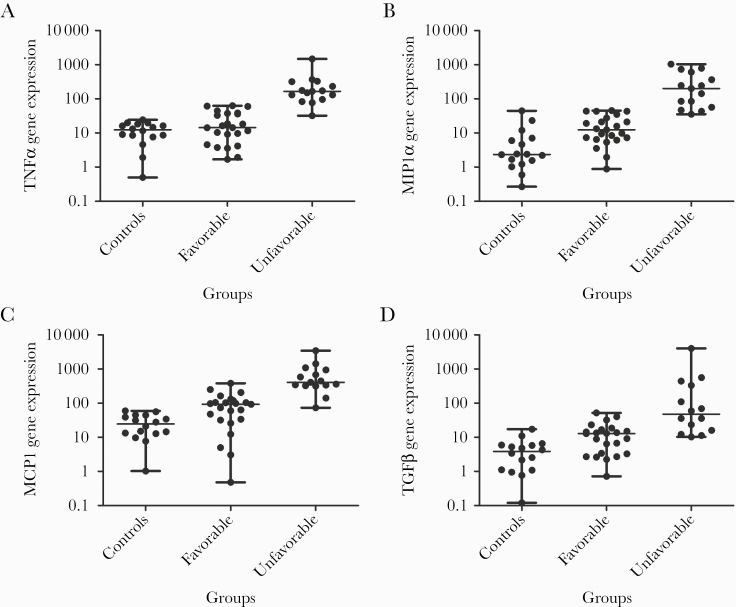
Figure 1A shows the comparison of TNFα expression among negative controls, patients with favorable and unfavorable evolutions. We did not find significant differences between the controls and patients with favorable evolution. The group with unfavorable evolution showed a significantly higher TNFα expression in comparison with the patients with a favorable evolution (P < .001) and with negative controls (P < .001).
Figure 1.
Relative gene expression of immune mediators in nasopharyngeal swab samples from coronavirus disease 2019 cases and controls. A, Expression of TNFα transcripts in different study groups of subjects: controls (samples from 16 healthy individuals confirmed as negative for severe acute respiratory syndrome coronavirus 2); favorable (23 subjects with favorable evolution: 12 asymptomatic subjects and 11 patients with mild disease); unfavorable (15 patients with unfavorable evolution: 9 very symptomatic patients and 6 who required intensive care; of these 6, 3 recovered and 3 died). B, Expression of CCL2/MCP-1 transcripts in those 3 groups. C, Expression of CCL3/MIP-1α transcripts in the 3 groups. D, Expression of TGFβ transcripts in the 3 groups. mRNA was determined for real-time reverse transcription polymerase chain reaction analysis, and the genes/housekeeping gene ratio was determined according to the delta/delta Ct method. Data are shown as dot plot graphics, on a log scale. Each dot plot shows statistic data about the gene expression determination. The central horizontal line in the dot marks the median of the samples; the hinges mark the ranges. Statistically significant differences in cytokine production among groups are indicated with horizontal lines over the dots. A P < .05 was considered statistically significant. P > .05 was considered not statistically significant. *P < .05; **P < .01; ***P < .001.
A significantly higher expression, however, of CCL2 was observed in cases with an unfavorable outcome compared with cases with a favorable evolution (P < .001) or with negative controls (P < .001) (Figure 1B). Similar results were obtained for CCL3: also here a significantly higher expression in cases with an unfavorable outcome was observed, compared with cases with a favorable evolution (P < .001) or with controls (P < .001) (Figure 1C).
The analysis of TGFβ expression among the 3 groups is shown in Figure 1D. Individuals grouped as having a favorable outcome showed a significantly higher expression compared with controls (P < .05). Patients with an unfavorable outcome showed higher expression than controls (P < .001) and cases with a favorable outcome (P < .01). In contrast, we did not find statistically relevant differences in IL-10 expression between individuals with favorable and unfavorable evolution or with negative controls (Supplementary Data).
We also analyzed if there were significant differences in the gene expression of each cytokine according to the time from symptom onset to sample collection. We found significantly higher expression only for TGFβ in the period from 0 to 3 days, compared with the period from 4 to 5 days (P = .005). We checked then which group of cases, according to clinical evolution, was supporting this difference in the period from 0 to 3 days, and we found significantly higher expression in cases with an unfavorable evolution compared with those with an favorable evolution (P = .033).
An association of TNFα with marked asthenia (P = .0004) and with the presence of dyspnea (P = .006) in the unfavorable evolution group was found. Dyspnea has been reported to be more frequent in severe COVID-19 cases, and indeed, in some studies, it has been included as a marker of severe disease [15, 16]. A similar association with these clinical symptoms was observed for CCL2/MCP-1 and TGFβ (asthenia: P = .003 and P = .006, respectively; dyspnea: P = .043 and P = .006, respectively) while CCL3/MIP-1α was associated only with asthenia (P = .004).
The univariate logistic regression models showed that the coefficients of 3 of the 5 cytokines evaluated were statistically associated with an unfavorable outcome (CCL2/MCP-1, TGFβ, and TNFα). Nevertheless, only CCL2/MCP-1 was significant in the “multivariate beta regression analysis (Estimate = 2.93; P = .024)”.
DISCUSSION
Early recognition of severe forms of COVID-19 is crucial for an opportune and effective early intervention that reduces life-risking complications.
A deregulated antiviral immune response, resulting in the release of large amounts of pro-inflammatory cytokines and subsequent uncontrolled local or systemic inflammation, has been recognized as causal for severity and lethality in pathogenic human coronavirus infections, including COVID-19 [3, 4].
As a consequence, several inflammatory markers, in particular cytokines, have been associated with severity and prognosis of COVID-19 [6]. However, most of these were detected in peripheral blood, and after the fifth day after onset of symptoms, which is relatively late [7].
To our knowledge, this is the first study to evaluate mRNA expression of cytokines/chemokines, already linked to COVID-19 pathogenesis, in swab samples from the upper airway at the early start of symptoms (first 5 days). Although the sample size was rather limited (16 healthy controls, 23 patients with favorable evolution, and 15 patients with unfavorable evolution), clearly statistical differences were observed.
Concurring with recent reports in Chinese COVID-19 patients, we found significantly higher expression of TNFα, CCL2/MCP-1, and CCL3/MIP-1α in COVID-19 cases with an unfavorable evolution compared with those with a favorable evolution [1, 8].
These markers have a high amplifying potential, quickly enhancing the inflammatory cytokine/chemokine responses in the upper airway, probably predicting later pathologic events in the lower airway associated to ARDS [9–11].
The higher levels of TGFβ in cases with the severest outcome could be in apparent contradiction with the regulatory and anti-inflammatory role attributed to this cytokine [12]. However, the function of TGFβ seems to be highly dependent on location and context and could play a major role in inflammatory conditions [13]; in fact, it has also been associated with hypoxia and lung damage [14]. The analysis of IL-10 gene expression did not show statistically relevant differences among the groups.
Cytokines are pleiotropic and interact as a network, in which the analysis of a single cytokine may not provide trustworthy information about serial immune events. Considering this, univariate logistic statistical analysis was conducted to detect differences in concentrations of each measured cytokine and to detect the confounders in patients with favorable vs unfavorable evolution. Also, in order to control for the effect confounders like age, sex, and the presence of comorbidities, a multivariate logistic regression analysis was used.
The univariate logistic regression model confirmed the association of TNFα, CCL2/MCP-1, and TGFβ with the COVID-19 outcome. However, after discarding the “noise” introduced by confounder variables, using a multivariate logistic regression model, only CCL2/MCP-1 was significant, which suggests the predictive value of this marker.
CCL2/MCP-1 regulates the migration and infiltration of monocytes, memory T lymphocytes, and natural killer (NK) cells, favoring inflammatory processes in tissues, including lung tissues [2]. Several recent reports have shown MCP-1 to be a biomarker predicting disease severity of COVID-19 in serum and plasma [1, 15, 16]. We report the predictive value of this chemokine in the mucosal environment, where the infection is occurring, and very early after symptom onset.
Moreover, the present study demonstrates the utility of nasopharyngeal swab samples collected within 5 days after the onset of symptoms for very early detection of immune markers. They may be a valuable and simple aid to improve the early identification of patients at risk for evolution toward a severe COVID-19 outcome, so that their health care providers can administer timely measures and treatments. Further research with a larger number of cases may validate CCL2/MCP-1 as an early mucosal biomarker predicting disease severity in COVID-19 patients.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Professor Xaveer Van Ostade, Laboratory for Protein Science, Proteomics and Epigenetic Signaling (PPES), Department of Biomedical Sciences (BMW), Faculty of Farmaceutical, Biomedical and Veterinary Sciences (FBD), University of Antwerp, for the revision of the manuscript.
Financial support. This work was supported by the Ministerio de Salud Pública de Cuba (MINSAP). Publishing charges for this article were supported by Vlaamse Interuniversitaire Raad-Universitaire Ontwikkelingssamenwerking (VLIR-UOS) through Project CU2019SIN243A102 (SI2019-SEL012).
Potential conflicts of interest. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu CM, Poon LL, Cheng VC, et al. Initial viral load and the outcomes of SARS. CMAJ 2004; 171:1349–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 2017; 39:529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mehta P, McAuley DF, Brown M, et al. ; HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 2020; 395:1033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. Laboratory testing strategy recommendations for COVID-19: interim guidance, 21 March 2020 2020. Available at: https://apps.who.int/iris/handle/10665/331509. Accessed 24 May 2020.
- 6. Zeng F, Huang Y, Guo Y, et al. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis 2020; 96:467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Velavan TP, Meyer CG. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis 2020; 95:304–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 2020; 9:761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 2009; 29:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zelova H, Hosek J. TNF-alpha signalling and inflammation: interactions between old acquaintances. Inflam Res 2013; 62:641–51. [DOI] [PubMed] [Google Scholar]
- 11. Glaser L, Coulter PJ, Shields M, et al. Airway epithelial derived cytokines and chemokines and their role in the immune response to respiratory syncytial virus infection. Pathogens 2019; 8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanjabi S, Zenewicz LA, Kamanaka M, Flavell RA. Anti-inflammatory and pro-inflammatory roles of TGF-beta, IL-10, and IL-22 in immunity and autoimmunity. Curr Opin Pharmacol 2009; 9:447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Påhlman LI, Jögi A, Gram M, et al. Hypoxia down-regulates expression of secretory leukocyte protease inhibitor in bronchial epithelial cells via TGF-β1. BMC Pulm Med 2015; 15:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosenbloom J, Macarak E, Piera-Velazquez S, Jimenez SA. Human fibrotic diseases: current challenges in fibrosis research. Methods Mol Biol 2017; 1627:1–23. [DOI] [PubMed] [Google Scholar]
- 15. Yang Y, Shen C, Li J, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol 2020; 146:119–27.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y, Wang J, Liu C, et al. IP-10 and MCP-1 as biomarkers predicting disease severity of COVID-19. [Preprint]. 2020. Available at: 10.21203/rs.3.rs-57499/v1. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.