Abstract
It has been widely accepted that mitochondria-dependent apoptosis initiates when select BH3-only proteins (BID, BIM, etc.) directly engage and allosterically activate effector proteins BAX/BAK. Here, through reconstitution of cells lacking all eight pro-apoptotic BH3-only proteins, we demonstrate that all BH3-only proteins primarily target the anti-apoptotic BCL-2 proteins BCL-xL/MCL-1, whose simultaneous suppression enables membrane-mediated spontaneous activation of BAX/BAK. BH3-only proteins’ apoptotic activities correlate with affinities for BCL-xL/MCL-1 instead of abilities to directly activate BAX/BAK. Further, BID and BIM do not distinguish BAX from BAK or accelerate BAX/BAK activation following inactivation of BCL-xL/MCL-1. Remarkably, death ligand-induced apoptosis in cells lacking BH3-only proteins and MCL-1 is fully restored by BID mutants capable of neutralizing BCL-xL, but not direct activation of BAX/BAK. Taken together, our findings provide a “Membrane-mediated Permissive” model, in which the BH3-only proteins only indirectly activate BAX/BAK by neutralizing the anti-apoptotic BCL-2 proteins, and thus allowing BAX/BAK to undergo unimpeded, spontaneous activation in the mitochondrial outer membrane milieu, leading to apoptosis initiation.
Subject terms: Apoptosis, Stress signalling
Introduction
The BCL-2 family proteins, which include the anti-apoptotic, the BH3-only, and BAX/BAK, are known to play critical roles in apoptosis signaling.1 In response to apoptotic stimulation, the BH3-only proteins suppress the anti-apoptotic BCL-2 proteins, and mediate the activation of effector proteins BAX and BAK.2 Activated BAX/BAK form homo-oligomers, and permeabilize the mitochondrial outer membrane (MOM), causing the release of cytochrome c and activation of caspases.3,4
Following two decades of intensive debate on the mechanism of BH3-only-mediated BAX/BAK activation, it has been widely accepted that the BH3-only proteins function in a hierarchy as either “sensitizers” or “direct activators” during apoptosis.5,6 In particular, the direct activators, e.g., BID, BIM, PUMA, etc., not only neutralize the anti-apoptotic BCL-2 proteins, but also use their BH3 domains to transiently engage BAX/BAK,7–10 triggering a conformational transformation of inactive BAX/BAK into active, oligomeric, and pore-forming molecules.11 On the other hand, BAD, a classic sensitizer that lacks the ability to bind and directly activate BAX/BAK, positively regulates apoptosis by binding to and neutralizing the anti-apoptotic BCL-2 proteins, namely BCL-2/BCL-xL/BCL-w,5,6,8 thus facilitating BAX/BAK activation by the direct activators. Using liposomes and in vitro reconstitution, truncated BID (tBID) and BIM have been shown to directly bind and activate BAX/BAK.5,10,12–14 Importantly, recent liposome-based studies have demonstrated that most pro-apoptotic BH3-only proteins, except for BAD, possess such ability.15–17 These results helped build a consensus that apoptosis initiates when BAX/BAK are directly and allosterically activated by the direct activator BH3-only proteins.18 However, as the list of reported “direct activator” BH3-only proteins has grown to include NOXA, BMF, BIK, and HRK,15–17,19 it has been extremely challenging to assess the role of “direct activation” during apoptosis initiation.
Other mechanisms of BAX/BAK activation have also been proposed over the years. Based on results from biochemical binding assays and BID/BIM double knockout (DKO) cells with a reduced expression of PUMA, Willis et al. proposed an Indirect (Displacement) model,20,21 in which BAX/BAK are postulated to be constitutively active but are normally sequestered by the anti-apoptotic BCL-2 proteins, and that the BH3-only proteins indirectly activate BAX/BAK by binding to the anti-apoptotic proteins, and thus releasing the active BAX/BAK. In another study, using elegant cell biology approaches, Edlich et al. presented data to suggest that while BAX has a natural tendency to move from the cytoplasm to the mitochondria, the anti-apoptotic BCL-2 protein BCL-xL possesses a retro-translocating activity that moves BAX from the mitochondria to the cytoplasm.22 However, due to the presence of multiple “direct activators” in any given cell, it remains unclear if direct activation plays a role in these two alternative mechanisms.
The generation of OctaKO cells,23 which lack all eight known pro-apoptotic BH3-only proteins, for the first time allows the examination of any given BH3-only protein or its mutants for their activities without participation of other BH3-only proteins. Furthermore, using cells deficient for the entire BCL-2 family, including the BH3-only, BAX/BAK, and pro-survival BCL-2 proteins,23 the role of BH3-only proteins after neutralization of the anti-apoptotic BCL-2 proteins can be investigated. In this study, by reconstituting the BH3-only-deficient and BCL-2 family-deficient cells, the contribution of “direct activation” in BH3-only-mediated BAX/BAK activation is directly assessed for the first time.
Results
BCL-xL and MCL-1 as direct targets of BH3-only proteins
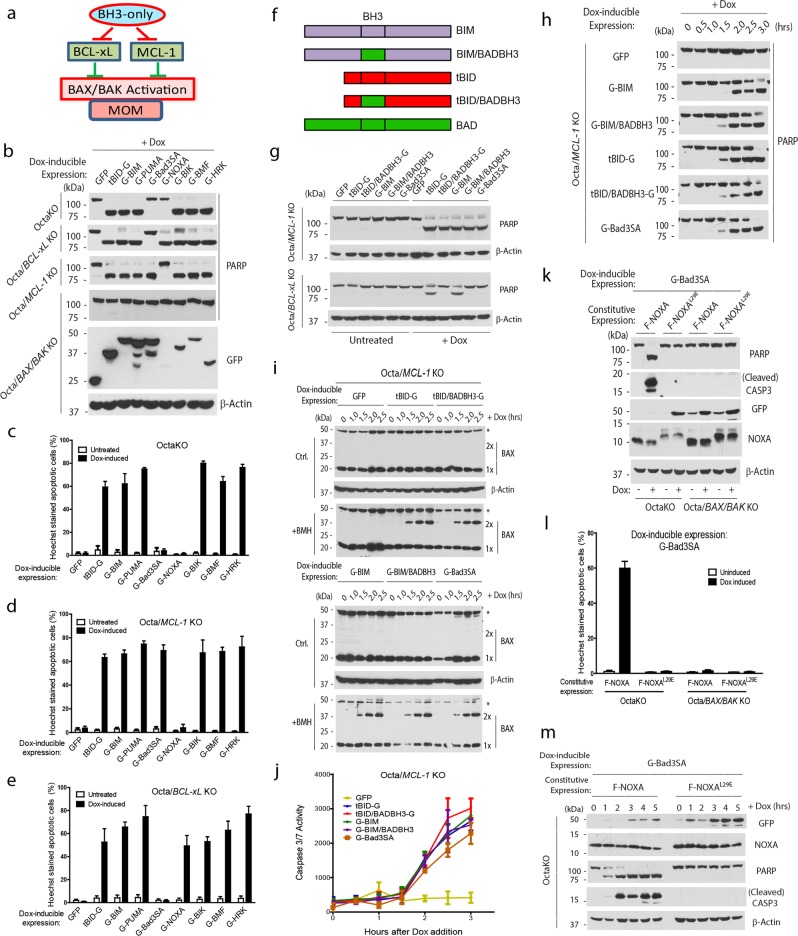
Concurrent elimination or inactivation of BCL-xL and MCL-1 has been found necessary and sufficient for BAX/BAK activation and apoptosis in the absence of all eight pro-apoptotic BH3-only proteins.23–25 This led us to hypothesize that neutralization of both BCL-xL and MCL-1 by the BH3-only proteins is sufficient for BAX/BAK activation (Fig. 1a). This hypothesis is tested by inducible expression of individual BH3-only proteins as GFP-fusion proteins in the OctaKO cells23 or their derivatives lacking either BCL-xL or MCL-1, and monitoring the progression of apoptosis by examining the cleavage of PARP (a caspase substrate) and CASPASE 3 (effector caspase), as well as chromosomal condensation or fragmentation (Hoechst staining). Of importance, the use of OctaKO-based cells should allow us to analyze the function of the BH3-only protein of interest without potential assistance from other BH3-only proteins. OctaKO, Octa/MCL-1 KO,23 Octa/BCL-xL KO (Supplementary information, Fig. S1 and Table S1), and Octa/BAX/BAK KO23 cells were therefore generated through CRISPR/Cas9, and infected with retrovirus to allow Dox-inducible expression of various BH3-only proteins. Following Dox addition, Bad3SA, an active mutant of BAD mimicking the dephosphorylated form of BAD, efficiently killed the Octa/MCL-1 KO cells, but failed to induce apoptosis in OctaKO or Octa/BCL-xL KO cells. On the other hand, NOXA killed only the Octa/BCL-xL KO cells, but not the OctaKO or Octa/MCL-1 KO cells. Of note, NOXA is difficult to detect due to its extremely short half-life.26 In contrast, tBID, BIM, PUMA, BIK, BMF, and HRK efficiently induced apoptosis in all three cell lines. As expected, none of the BH3-only proteins displayed any apoptotic activity in the Octa/BAX/BAK KO cells, indicating a BAX/BAK dependency of BH3-only protein-induced apoptosis (Fig. 1b–e; Supplementary information, Fig. S2a). These results strongly suggest that BAD and NOXA target BCL-xL and MCL-1, respectively, and that tBID, BIM, PUMA, HRK, BMF, and BIK target both BCL-xL and MCL-1 during apoptotic signaling. This conclusion is consistent with the differential affinities between BH3-only and anti-apoptotic BCL-2 proteins as defined through in vitro binding assays.27 As BAD is known to only bind BCL-2, BCL-xL, BCL-w, but not BAX/BAK, the robust apoptotic activity of Bad3SA in Octa/MCL-1 KO cells strongly suggests that neutralization of BCL-xL and MCL-1 is sufficient for BAX/BAK activation.
Fig. 1.
All BH3-only proteins can trigger BAX/BAK activation by targeting only BCL-xL/MCL-1. a Hypothesis of BH3-only-mediated BAX/BAK activation. b Dox-inducible expression of the BH3-only-GFP-fusion proteins was established in the listed four parental HCT116 knockout cell lines by puromycin selection. GFP is at the N-termini of most of the BH3-only proteins except for tBID, whose BH3 domain is at the N-terminus. Dox (2 µg/mL) was added to each of the selected pools expressing different BH3-only proteins as indicated. Eight hours after induction, cells were harvested for western blot analysis. The faster migrating protein species in lane 4 and 5 for the Octa/BAX/BAK KO cells are minor degradation products of G-PUMA and G-Bad3SA. c–e Cells in b were stained with Hoechst dye 8 h after Dox induction. The percentage of cells that underwent nuclear condensation or fragmentation was quantified. The results are averages with SEM of three independent experiments. f Diagram of the listed BH3-only proteins and their BADBH3 mutants. g Dox-inducible expression for the listed GFP or GFP-fusion proteins were established in Octa/MCL-1 KO and Octa/BCL-xL KO HCT116 cells. Cells were treated with Dox (2 µg/mL) for 3 h and were harvested for western blot analysis. h At the indicated time points after Dox addition, cells were harvested for western blot analysis. i Cells were treated with Dox (2 µg/mL) in the presence of z-VAD for the indicated amount of time before being harvested. Whole cell lysates were incubated with or without BMH (0.2 mM) at room temperature for 1 h. The reaction mixtures were then subjected to western blot analysis. j Cells in h were induced with Dox for the indicated amount of time. Apo-ONE Caspase substrate/buffer for Caspase 3/7 were added to cells at the end of the treatment, and the plates were analyzed on a fluorescence microplate reader. The results are averages with SEM of three independent experiments. k Stable and Dox-inducible expression of the indicated proteins were established in the listed knockout cell lines. Four hours after Dox addition, cells were harvested for western blot analysis. l Cells in k were stained with Hoechst dye 4 h after Dox induction. The percentage of cells that underwent nuclear condensation or fragmentation was quantified. The results are averages with SEM of three independent experiments. m Stable and Dox-inducible expression of the indicated proteins were established in the OctaKO cells. At different time points after the addition of Dox, cells were harvested for western blot analysis. Representatives from at least two independent experiments are shown for western blots
To further examine the role of direct activation in BAX/BAK activation, we sought to compare the apoptotic kinetics of the two classic direct activators, tBID and BIM, which bind BAX/BAK, BCL-xL and MCL-1, with that of the classic sensitizer, BAD (Bad3SA), which binds BCL-xL, BCL-2, and BCL-w, but not BAX/BAK or MCL-1,27 in the absence of other BH3-only proteins. Since all three proteins bind BCL-xL, we chose to conduct the kinetics comparison in Octa/MCL-1 KO cells, whose survival is strictly dependent on endogenous BCL-xL. We reasoned that if the ability to bind BAX/BAK plays a role in BAX/BAK activation, tBID and BIM should display faster kinetics as compared to Bad3SA, as the latter is unable to bind BAX/BAK. As shown in Fig. 1g–j and Supplementary information, Fig. S2c–g, however, Bad3SA induced a robust BAX/BAK activation with similar kinetics to that of tBID and BIM, indicating that tBID and BIM’s putative direct activation activities are not necessary for BAX/BAK activation, and that such activities do not have discernable effects on BAX/BAK activation following neutralization of BCL-xL.
However, it remains possible that the direct activator BH3-only proteins, e.g., BID and BIM, use both direct and indirect mechanisms to activate BAX/BAK. To examine the contribution of direct activation and neutralization-mediated indirect activation in BAX/BAK activation by tBID or BIM, we replaced the BH3 domains of tBID and BIM with that of BAD (Fig. 1f; Supplementary information, Fig. S2b). Since the BH3 domain of BAD (BADBH3) is known to only bind BCL-2/BCL-xL/BCL-w,27 such mutants should retain the ability to neutralize BCL-xL, but not Mcl-1, and it is expected to completely lose the ability of “direct activation”. When tested in Octa/BCL-xL KO cells, unlike wild-type tBID and BIM, tBID/BADBH3, BIM/BADBH3, and Bad3SA are unable to induce apoptosis, indicating that the binding specificities of these tBID and BIM BH3 mutants are indeed converted to that of BAD (Fig. 1g; Supplementary information 2c). In contrast, when expressed in the Octa/MCL-1 KO cells, both BH3 mutants of tBID and BIM, as well as Bad3SA displayed apoptotic kinetics indistinguishable from those of wild-type tBID and BIM (Fig. 1h, j; Supplementary information, Fig. S2d, e). As BAD has been widely accepted as a protein lacking direct activation activity, and that tBID/BADBH3 was shown to be completely unable to directly activate BAX in liposome assays,16 these results indicate that the putative “direct activation” activities of these “direct activators” do not contribute to their ability to activate BAX/BAX.
To more directly monitor BAX activation induced by the wild-type and mutants of the above mentioned BH3-only proteins, we devised a BAX dimerization/oligomerization assay to monitor the kinetics of BAX dimer formation, a well-recognized event leading to BAX activation following apoptotic stimulation. In this assay, cell extracts are prepared at different time points after the induction of the BH3-only or BAX proteins. Following incubation with the cross-linker BMH, the cell extracts are examined for the formation of dimers of either the endogenous or induced BAX by western blot. The 40-kD protein species is confirmed to be BAX dimers as Octa/MCL-1/BAX KO cells failed to generate the 40-kD protein in the BMH assay following tBid induction (Supplementary information, Fig. S2f). As shown in Fig. 1i and Supplementary information Fig. S2g, in Octa/MCL-1 KO cells, wild-type tBID, BIM, their BADBH3 mutants, and Bad3SA, but not GFP, induced endogenous BAX dimer formation and apoptosis with almost identical kinetics, strongly suggesting that neutralization of the anti-apoptotic BCL-2 proteins is sufficient for BAX/BAK activation, and that direct activation by the “direct activators” contributes little, if any, to BAX/BAK activation.
Considering the selective binding of BAD and NOXA to BCL-xL and MCL-1, respectively,27 we examined the cooperativity of these two proteins in inducing apoptosis in the OctaKO cells (Fig. 1b). Retroviral infection was used to stably express NOXA or its BH3 defective mutant NOXAL29E in OctaKO cells. A Dox-inducible system was generated for the expression of Bad3SA. As shown in Fig. 1k–m, the Dox-induced expression of Bad3SA induced robust apoptosis in the presence of NOXA, but not NOXAL29E, suggesting that neutralization of both BCL-xL and MCL-1 by individual BH3-only proteins or their combinations is sufficient to trigger BAX/BAK activation.
BIM and BID activate BAX/BAK without preference for BAX or BAK
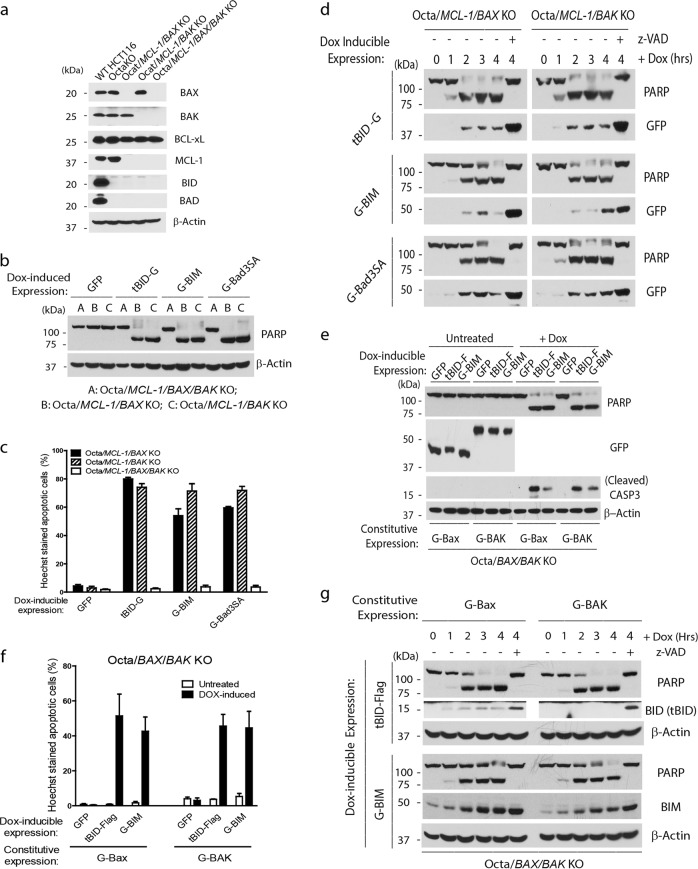
In support of the “Direct Activation” model, data from an in vitro liposome reconstitution study suggested that the two classic direct activators, tBID and BIM, possess differential activities towards BAX and BAK, with tBID preferentially activating BAK, and BIM preferentially activating BAX.14 We therefore sought to examine such preferential activation through reconstitution in the OctaKO derivative cells that are deficient for either BAX or BAK (Fig. 2a; Supplementary information, Fig. S3 and Table S1). In addition, MCL-1 was eliminated so that BAD can be functionally examined in the reconstitution study (Fig. 2a; Supplementary information, Table S1). In both Octa/MCL-1/BAX KO and Octa/MCL-1/BAK KO cells, we established the Dox-inducible system for the expression of tBID, BIM, or Bad3SA (Supplementary information, Fig. S4a). Following Dox addition, all three proteins caused robust apoptosis in both cell lines. As expected, none of these proteins induced apoptosis in the Octa/MCL-1/BAX/BAK KO cells (Fig. 2b, c). Importantly, upon Dox-induced expression, all three proteins induced apoptosis with similar kinetics in the presence of either endogenous BAX or Bak (Fig. 2d; Supplementary information, Fig. S4b, c).
Fig. 2.
BIM, tBID, and BAD do not distinguish BAX from BAK. a Generation of Octa/MCL-1/BAX KO and Octa/MCL-1/BAK KO cells. Whole cell lysates for the indicated cells were analyzed by western blot analysis. b Dox-inducible expression for the indicated proteins was established in Octa/MCL-1/BAX/BAK KO, Octa/MCL-1/BAX KO, and Octa/MCL-1/BAK KO cells. Four hours after Dox (2 µg/mL) addition, cells were harvested for western blot analysis. c Cells were treated the same way as in b and were stained with Hoechst dye after induction. The percentage of cells that underwent nuclear condensation or fragmentation was quantified. The results are averages with SEM of three independent experiments. d The Octa/MCL-1/BAX KO and Octa/MCL-1/BAK KO cells with the Dox-induced expression of the indicated proteins were harvested at different time points following the addition of Dox for western blot analysis. e Dox-inducible expression of the indicated proteins was established in Octa/BAX/BAK KO cells stably expressing comparable amounts of G-Bax or G-BAK. Four hours after induction, cells were harvested for western blot analysis. f Cells were treated the same way as in e and then stained with Hoechst dye. The percentage of cells that underwent nuclear condensation or fragmentation was quantified. The results are averages with SEM of three independent experiments. g Cells in e were harvested at the indicated time points after Dox addition for western blot analysis. Representatives from at least two independent experiments are shown for western blots
As the above experiments rely on endogenous BAX or BAK, the relative levels of BAX and BAK in the cells may affect the kinetics of apoptosis. We therefore reconstituted the Octa/BAX/BAK KO cells with either GFP-Bax or GFP-BAK at comparable levels (Fig. 2e). In both settings, tBID and BIM induced apoptosis with similar kinetics (Fig. 2e–g; Supplementary information, Fig. S4d, e). These results are inconsistent with the notion that tBID and BIM preferentially activate BAK or BAX, respectively.14 Using BID chimeric proteins in liposome assays, Hockings et al. was also not able to observe a preference for BAX or BAK by the BH3 domains.16 Together, these results support the notion that tBID and BIM’s primary targets are the anti-apoptotic BCL-2 family proteins, whose neutralization is sufficient to allow spontaneous BAX/BAK activation.
Role of BH3-only proteins after neutralization of the anti-apoptotic BCL-2 proteins
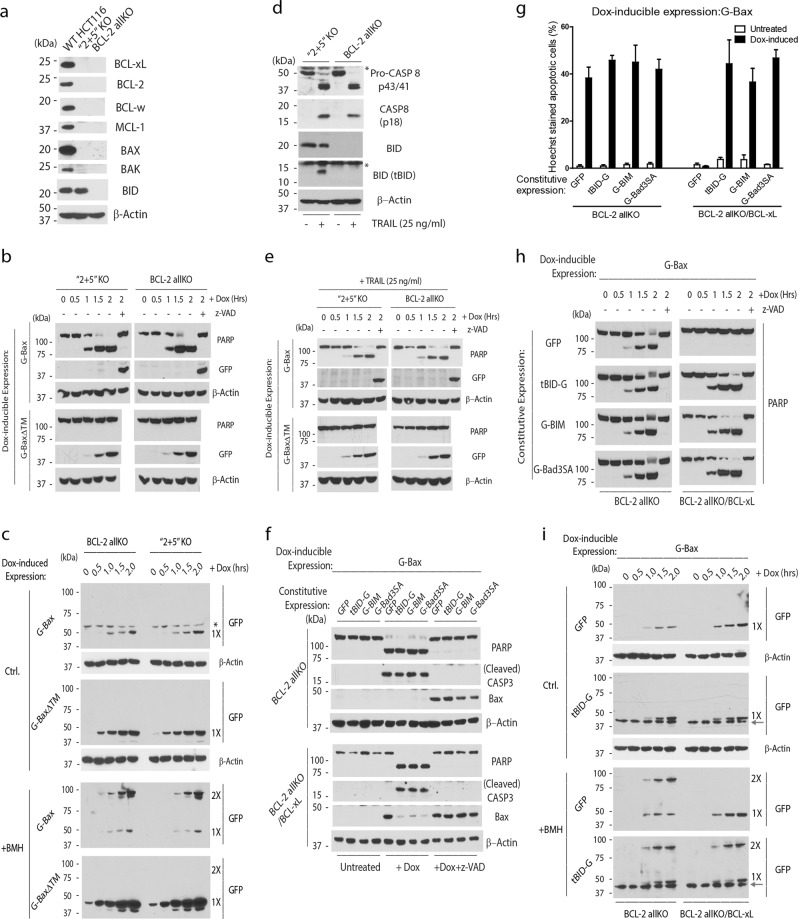
Although the above results suggest that direct activation activity is not necessary for BAX/BAK activation, it remains possible that after neutralizing the anti-apoptotic BCL-2 proteins, the direct activators interact with free BAX/BAK, and accelerate their membrane-mediated spontaneous activation. We therefore sought to examine the role of BH3-only proteins in activating BAX under the cellular background where all anti-apoptotic BCL-2 proteins are eliminated. To achieve this, we used CRISPR/Cas9 to generate “2 + 5” cells, which are deficient for BAX/BAK and all five major anti-apoptotic BCL-2 family proteins (Fig. 3a; Supplementary information, Fig. S5 and Table S1). The lack of anti-apoptotic BCL-2 proteins is expected to render most of the BH3-only proteins constitutively active. A Dox-inducible system for the inducible expression of G-Bax was established in both the “2 + 5” cells and the BCL-2 allKO cells, which are deficient for the 17 BCL-2 family members, including the BH3-only proteins, BAX, BAK, and the five anti-apoptotic BCL-2 proteins.23 As shown in Fig. 3b, c and Supplementary information, Fig. S6a, b, the kinetics of Dox-induced Bax activation (dimer formation) and apoptosis, as measured in the BMH assay and by PARP cleavage, are similar in both BCL-2 allKO and “2 + 5” cells, indicating that the endogenous BH3-only proteins do not contribute to BAX/BAK activation in the absence of the anti-apoptotic BCL-2 proteins. This spontaneous activation is entirely mediated by mitochondrial membrane association, as G-BaxΔTM completely lost the capacity to induce apoptosis in either “2 + 5” or BCL-2 allKO cells (Fig. 3b, c; Supplementary information, Fig. S6b). Further, we treated both cells with the death ligand TRAIL, generating tBID through the death receptor pathway in the “2 + 5” cells (Fig. 3d). Similarly, the presence of tBID in the “2 + 5” cells did not accelerate the Dox-induced spontaneous BAX activation, which is completely abolished for the Bax mutant that lacks the mitochondrial outer membrane targeting sequence (Fig. 3e; Supplementary information, Fig. S6c, d).
Fig. 3.
BIM and tBID inactivate BCL-xL/MCL-1, but do not accelerate BAX/BAK activation in the absence of the anti-apoptotic BCL-2 proteins. a BAX/BAK/BCL-xL/BCL-2/BCL-w/MCL-1/A1 KO cells, which are named “2 + 5” cells, were generated with CRISPR/Cas9. Whole cell lysates were examined for the expression of the indicated proteins by western blot analysis. b Dox-inducible expression of G-Bax or G-BaxΔTM was established. At the indicated time points, cells were harvested for western blot analysis. c Cells were treated with Dox in the presence of z-VAD for the indicated amount of time before harvested. Whole cell lysates were incubated with or without BMH (0.2 mM) at room temperature for 1 h. The reaction mixtures were then subjected to western blot analysis. An asterisk indicates a non-specific protein. d The indicated cells were treated with TRAIL (25 ng/mL) for 3 h before they were harvested for western blot analysis. An asterisk indicates a non-specific protein. e Three hours after TRAIL treatment, Dox was added to the indicated cell lines with the Dox-inducible expression of either G-Bax or G-BaxΔTM. Cells were harvested at different time points after addition of Dox for western blot analysis. f Dox-inducible expression system for G-Bax was established in BCL-2 allKO cells expressing the indicated BH3-only proteins as GFP chimeras with or without the stable expression of BCL-xL. Dox (2 µg/mL) was added in the presence or absence of z-VAD (40 µM) for 2 h before the cells were harvested for western blot analysis. g Cells in f were stained by Hoechst dye 2 h after Dox induction. The percentage of cells that underwent nuclear condensation or fragmentation was quantified. The results are averages with SEM of three independent experiments. h At the indicated time points following the addition of Dox, cells from f with the expression of the indicated proteins were harvested for western blot analysis with PARP antibody. i Cells were treated with Dox in the presence of z-VAD for the indicated amount of time before harvested. Whole cell lysates were incubated with or without BMH at room temperature for 1 h. The reaction mixtures were then subjected to western blot analysis. Arrows indicate the protein bands of tBID-G. Representatives from at least two independent experiments are shown for western blots. An asterisk indicates a non-specific protein
To further examine the effect of tBID, BIM, or BAD on BAX activation after neutralization of the anti-apoptotic proteins, we stably expressed GFP-tagged BH3-only proteins in the BCL-2 allKO cells with or without stable expression of BCL-xL (Supplementary information, Fig. S6e). It is worth mentioning that in the “2 + 5” and BCL-2 allKO cells, due to the lack of the anti-apoptotic BCL-2 proteins, the only target of the ectopically expressed BH3-only proteins is Dox-induced Bax according to the “Direct Activation” model. In the absence of BCL-xL, Dox-induced GFP-Bax and its spontaneous activation caused apoptosis with similar kinetics in cells expressing GFP, tBID-GFP, G-BIM, or G-Bad3SA. On the other hand, in the presence of BCL-xL, Dox-induced G-Bax failed to spontaneously activate in the BCL-2 allKO cells, indicating that BCL-xL is capable of inhibiting the spontaneous BAX activation (Fig. 3f–i; Supplementary information, Fig. S6f–i). However, the expression of tBID-GFP, G-BIM or G-Bad3SA completely overcame such inhibition, indicating that the expressed tBID, BIM, and Bad3SA are indeed active (Fig. 3f–i; Supplementary information, Fig. S6f–i). These results strongly suggest that the main function of the BH3-only proteins is to suppress the anti-apoptotic BCL-2 proteins, whose function is to suppress the spontaneous activation of BAX/BAK. Thus, once the anti-apoptotic BCL-2 proteins are neutralized, Bax undergoes mitochondrial outer membrane-mediated activation without the need for the BH3-only proteins.
Reconstitution of death receptor-mediated, mitochondria-dependent apoptosis pathway by BID or its mutants
To examine the role of “direct activation” activity of direct activator BH3-only proteins during apoptotic signaling, we resorted to the cell surface death receptor-mediated pathway. Following death receptor-mediated CASPASE 8 activation, BID is cleaved by CASPASE 8 to give rise to tBID, which then moves to the mitochondria to cause BAX/BAK activation.28,29 We therefore reconstituted the OctaKO and Octa/MCL-1 KO cells with endogenous levels of wild-type BID or several of its mutants with altered BH3 domains through retroviral expression (Fig. 4a, b). The first mutant, BIDI82A/I83A, was found to be defective in direct activation in an in vitro liposome assay.7 A second mutant, BID/BADBH3, has its BH3 domain replaced by that of the sensitizer protein BAD.16 BIDG94E, a BH3 mutant known to be defective in binding to BCL-2/BCL-xL,29 was used as a negative control. As expected, while cells reconstituted with GFP or BIDG94E failed to undergo apoptosis in response to TRAIL, expression of wild-type BID restored apoptosis, indicating the requirement of a functional BID in this pathway (Fig. 4c–h; Supplementary information, Fig. S7a, b). Remarkably, expression of BIDI82A/I83A also fully restored apoptosis in response to TRAIL in both OctaKO and Octa/MCL-1 KO cells (Fig. 4c–h; Supplementary information, Fig. S7a, b). In contrast, BID/BADBH3 failed to restore apoptosis in OctaKO cells (Fig. 4c, d, f, g; Supplementary information, Fig. S7a, b). However, such failure appears to be due to BID/BADBH3’s inability to bind to and neutralize endogenous MCL-1, as addition of S63845,30 a highly selective MCL-1 inhibitor, to OctaKO cells restored the response to TRAIL (Fig. 4c, d; Supplementary information, Fig. S7a). Consistently, BID/BADBH3 completely restored apoptosis to Octa/MCL-1 KO cells in response to TRAIL treatment (Fig. 4c, e, f, h; Supplementary information, Fig. S7a, b). Thus, TRAIL-mediated apoptosis pathway can be fully reconstituted by BID mutants capable of neutralizing the anti-apoptotic BCL-2 proteins but defective in direct activation, strongly suggesting that death ligand-induced, BAX/BAK-dependent apoptotic pathway is mediated by the neutralization activity of the BH3-only protein BID against BCL-xL and MCL-1, following BID’s activation by CASPASE 8-mediated cleavage and association with the mitochondrial outer membrane (Fig. 4i).31
Fig. 4.
Reconstitution of the death receptor-mediated, BAX/BAK-dependent apoptosis pathway with BID and its direct activation-defective mutants in BH3-only-deficient cells. a Diagram of BID and its mutants. b Stable expression of BID and its mutants in OctaKO and Octa/MCL-1 KO cells following retroviral infection and flow cytometry sorting. c The indicated stable cell pools were treated with TRAIL (25 ng/mL) in the presence or absence of S63845 (1 µM) for 2.5 h and then harvested for western blot analysis. d, e Cells in c were stained by Hoechst dye 2.5 h after Dox induction. The percentage of cells that underwent nuclear condensation or fragmentation was quantified. The results are averages with SEM of three independent experiments. f The indicated stable cell pools were treated with TRAIL (25 ng/mL) for the indicated amounts of time and then harvested for western blot analysis. g, h Cells were treated with TRAIL (25 ng/mL) for the indicated amount of time. Apo-ONE Caspase substrate/buffer for Caspase 3/7 were added to cells at the end of the treatments, and the plates were analyzed on a fluorescence microplate reader. The results are averages with SEM of three independent experiments. i Diagram of the death receptor-mediated, mitochondria-dependent apoptosis pathway. Representatives from at least two independent experiments are shown for western blots
Discussion
The mechanism of BAX/BAK activation has been a major topic of debate in the past two decades.32 The “Direct Activation” model5,6,9,10,17,33 and the closely related “Unified” model,34 both of which gained wide recognition, suggest that a subgroup of the BH3-only proteins, for example, BID and BIM, not only target the anti-apoptotic BCL-2 proteins but also BAX/BAK, and such direct BH3-only-BAX/BAK interaction is the trigger or catalyst for BAX/BAK activation.34–36 Due to the large number of BH3-only proteins with overlapping functions, the physiological significance of this model has been difficult to assess.37 The current study eliminated the possibility of assistance from other BH3-only proteins, and directly addressed the functional contribution of the “direct activation” activity of select BH3-only proteins during apoptosis.
BIM and tBID, two most recognized “direct activators”, and BAD, a classic “sensitizer”, are functionally compared for the first time in BH3-only-deficient cells that also lack MCL-1 (Octa/MCL-1 KO) (Figs. 1–3). Such comparison provides answers to two important questions. First, is neutralization of anti-apoptotic BCL-2 proteins sufficient for BAX/BAK activation? Second, what are the contributions of neutralization and direct activation, two separate activities, to the ability of a direct activator BH3-only protein to activate BAX/BAK? Especially, the requirement of tBID’s direct activation activity was examined in the context of TRAIL-induced apoptosis (Fig. 4). To our surprise, under simplified genetic backgrounds, the “sensitizers” (i.e., Bad) and the direct activation-incompetent mutants of “direct activators” (i.e., tBID/BADBH3) displayed apoptotic activities indistinguishable from those of the “direct activators”, strongly suggesting that the putative “direct activation” activity does not contribute to BAX/BAK activation, and that the neutralization activity of BH3-only proteins is sufficient. Although it remains possible that some BH3-only proteins transiently bind free BAX or BAK as suggested by results from the in vitro studies, data obtained from anti-apoptotic BCL-2-deficient cells (Fig. 3) suggests that such interactions are non-essential, and unlikely to serve a functional role in BAX/BAK activation. On the other hand, the free diffusion-based, spontaneous, and high-affinity binding of BAX/BAK to the MOM, most likely the lipids in the MOM, is required and sufficient for BAX/BAK activation (Fig. 3). Consistent with an active role of the lipids in BAX/BAK activation, it has been shown that BAX/BAK are efficiently activated by non-ionic detergents in vitro.38,39
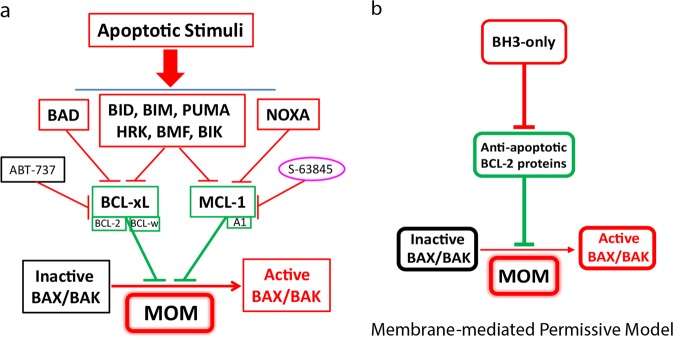
The current study for the first time unequivocally defined the functional relationships among the BCL-2 family proteins. Unlike the “Direct Activation” or “Unified” models,5,17,34 which suggest that BH3-only proteins and anti-apoptotic BCL-2 proteins are mutual inhibitors (Supplementary information, Fig. S8), our data clearly suggests unidirectional relationships in which the BH3-only proteins inhibit anti-apoptotic BCL-2 proteins, whose major function is to inhibit membrane-mediated BAX/BAK activation (Fig. 5). This conclusion is consistent with the original “Indirect Activation/Displacement” model in which BH3-only proteins inactivate anti-apoptotic BCL-2 proteins but do not directly bind/activate BAX/BAK (Supplementary information, Fig. S8).21 However, key differences exist. The latter model relies on the notion that BAX/BAK are constitutively active but are sequestered by the anti-apoptotic BCL-2 proteins before apoptosis, and therefore suggests that the BH3-only proteins initiate apoptosis by binding the anti-apoptotic BCL-2 proteins, thus displacing and releasing active BAX/BAK (Supplementary information, Fig. S8).20,40 Such a scenario has difficulty explaining the apparent differential localization of BAX and BCL-2/xL proteins in healthy cells. In contrast, in combination with results from a previous study,23 our data strongly supports a novel “Membrane-mediated permissive” model (Fig. 5), which suggests that the vast majority, if not all, of BAX/BAK molecules exists as inactive monomers (BAX) or homo-dimers (BAK) before apoptosis,7,39 but is uniformly activated through their spontaneous, free diffusion-driven association with the MOM, once the anti-apoptotic BCL-2 proteins are neutralized by the BH3-only proteins or the BH3 mimetics30,41 during apoptosis initiation (Fig. 5). Consistent with this model, Greaves et al. demonstrated that pharmacological inhibition of BCL-xL and MCL-1 efficiently induces apoptosis in the absence of the BH3-only proteins.42 Moreover, the observation that BAX has a natural tendency of translocating to the mitochondria, but can be retro-translocated back into the cytosol by BCL-xL is consistent with this model.22
Fig. 5.
A new model of BAX/BAK activation during apoptosis. a BH3-only-mediated BAX/BAK activation during mitochondria-dependent apoptotic signaling. In response to apoptotic signals, select groups of BH3-only proteins become activated, and the activated BH3-only proteins neutralize the anti-apoptotic BCL-2 proteins, allowing the free diffusion-driven, membrane-mediated, spontaneous activation of BAX/BAK through the mitochondrial outer membrane. While NOXA binds only MCL-1 and A1, and BAD binds only BCL-xL, BCL-2, and BCL-w, BID, BIM, PUMA, HRK, BMF, and BIK are able to bind all the anti-apoptotic BCL-2 proteins. b A mitochondrial outer membrane-mediated permissive model of BAX/BAK activation. BAX/BAK are normally inactive monomers or dimers in healthy cells. During apoptosis, activated BH3-only proteins bind to and neutralize the anti-apoptotic BCL-2 family proteins on the mitochondrial outer membrane, which then provides a permissive environment for the spontaneous activation of BAX/BAK, culminating in their homo-oligomerization and the formation of cytochrome c releasing pores
Overall, our results strongly support the notion that the BH3-only proteins activate BAX/BAK primarily by targeting and neutralizing the anti-apoptotic BCL-2 proteins, therefore allowing BAX/BAK’s spontaneous activation in the mitochondrial outer membrane.23 This new model of BAX/BAK activation, therefore, largely excludes any role of direct activation by the “direct activator” BH3-only proteins once the anti-apoptotic BCL-2 proteins are neutralized. In other words, the “direct activator” BH3-only proteins (BID, BIM, etc.) are similar to the “sensitizers” (i.e., BAD), because their only function is to neutralize the anti-apoptotic BCL-2 proteins. To that end, although OctaKO and BCL-2 allKO cells provide the necessary genetic background to understand the seemingly complex functional relationship among the BCL-2 family proteins, it is worth pointing out that our conclusions are based on genetic experiments carried out in HCT116 cells. Theoretically, it remains possible that in other cellular contexts, direct activation may play a positive role in BAX/BAK activation. However, such a role may only be revealed when it is examined in a cellular environment without the interference and complication from other BH3-only proteins.
Materials and methods
Cell culture
HCT116 cells (ATCC) were cultured in McCoy’s 5A medium with penicillin/ streptomycin and 10% fetal bovine serum (Atlanta Biologicals, #S11150). 293GP cells and 293T cells were grown in DMEM media with penicillin/streptomycin and 10% FBS. All cells were maintained at 37 °C with 5% CO2.
Reagents
Doxycycline Hydrochloride was purchased from Fisher Scientific. Human recombinant TRAIL was generated as previously described43. z-VAD-FMK was purchased from MP Biomedicals. S63845 was purchased from ChemieTek (IN, USA). Antibodies used include anti-PARP (Cell Signaling Technologies, no. 9524), anti-GFP (Santa Cruz Biotechnology, no. sc-459), anti-Flag (Sigma-Aldrich, no. F1804), anti-BCL-xL (Santa Cruz Biotechnology, no. sc-8392), anti-BCL-2 (Santa Cruz Biotechnology, no. sc-509), anti-MCL-1 (Santa Cruz Biotechnology, no. sc-819), anti-BIM (Calbiochem, #202000), anti-BID,29 anti-BAX (Cell Signaling Technologies, no. 2772), anti-BAK (Santa Cruz Biotechnology, no. sc-832), anti-BCL-w (Cell Signaling Technologies, no. 2724), anti-BAD (Santa Cruz Biotechnology, no. sc-8044), anti-NOXA (Santa Cruz Biotechnology, no. sc-56169) anti-Tet-R (Clontech, no. 631131), anti-CASPASE 8 (Cell Signaling Technologies, no. 9746), anti-CASPASE 3 (Cell Signaling Technologies, no. 9665S) and anti-β-actin (Sigma-Aldrich, no. A5441).
Immunoblotting
Cells were harvested and lysed in EBC buffer (120 mM NaCl, 50 mM Tris, 1 mM EDTA, 0.5% NP40, pH = 7.5) with protease inhibitors (5 mg/mL pepstatin A, 10 mg/mL leupeptin) and 0.1 mM PMSF. After rotating for 1–2 h at 4 °C and centrifugation at 22,000 × g for 10 min at 4 °C, supernatant was collected as the whole cell lysate. Protein concentrations were measured by Coomassie Protein Assay solution (Thermo Scientific, # 1856209). Cell lysate was mixed with 4× SDS loading buffer and heated at 98 °C for 5 min before resolved by SDS-PAGE gel and transferred onto a nitrocellulose membrane, which was then incubated with primary and secondary antibodies for western blotting. Proteins of interest were detected with Chemiluminescence.
Plasmids
CRISPR plasmids used were generated as described before.23 The retroviral vector pRxXE-9HG was generated by ligating a fragment containing 9 × His-GFP and XhoI cut site into the retroviral vector pRetroX-Tight-Pur vector (Clontech) and site directed mutagenesis to disable the XhoI site preexisted in the vector. Human tBID, BIM, PUMA, BIK, BMF, HRK, NOXA, and mouse Bad3SA were cut from respective pEGFP-C3 constructs and ligated into the pRxXE-9HG vector with XhoI and EcoRI sites.
BIM, BID, and tBID mutant with BH3 domain replaced with BAD BH3 (BIM/BADBH3, BID/BADBH3, and tBID/BADBH3) was generated by PCR SOEing. Human BIM R143-Y162 and human BID D81- S100 were replaced by human BAD W105- S124. The cDNA for BIM/BADBH3 was cloned into pRxXE-9HG vector with XhoI and EcoRI sites. The cDNA for tBID-GFP or tBID/BADBH3-GFP was cloned into pRetroX-Tight-Pur vector with NotI and EcoRI sites.
The retroviral expression plasmids pMIG-GFP and pMIG-BCL-xL were generated as described before.23 Flag-NOXA and Flag-NOXA L29E were PCR amplified from respective pcFlag constructs and cloned into MSCV-IRES-GFP (pMIG) vector using XhoI and HindIII sites. GFP-Bax, GFP-BAK, GFP-BIM, tBID-GFP and GFP-Bad3SA were PCR amplified from respective pEGFP-C3 or pEGFP-N2 constructs and cloned into MSCV-IRES-GFP (pMIG) vector using EcoRI and HindIII sites. Tetracycline repressor gene was PCR amplified from pRetroX-Tet-on Advanced plasmid and cloned into mpMIG (modified pMIG) using XhoI and EcoRI sites. GFP-Bax and GFP-BaxΔTM were PCR amplified and cloned into lentiviral vector pCW57.1 (Addgene #41393) with EcoRI and MluI sites. BIDI82A/I83A and BIDG94E were generated by standard site directed mutagenesis. BID, BID/BADBH3, BIDI82A/I83A and BIDG94E were cloned into mpMIG (modified from pMIG) vector with XhoI and EcoRI sites.
Plasmid transfection
For CRISPR transfection, cells were seeded in a 35 mm plate at the density of 3–4 × 105 cells per plate 24 h prior to transfections. Five hundred nanograms of each CRISPR plasmid was co-transfected with 300 ng of the corresponding mRFP-TS-2A-HYG-EGFP reporter plasmid. pcDNA3.1 was added to make the total DNA concentration of 1.8 µg per transfection. Twenty four hours after transfection, the transfected cells were split 1:2 and subjected to flow cytometry sorting for GFP/RFP-positive cells or Hygromycin B (1 mg/mL, Calbiochem, #400050) selection. The sorted or selected cells were plated for single clones in 15 cm plates.
Flow cytometry
BD FACSAria or a BD FACSAria II cell sorter with BD FACSDiva 6.1.2 software (UNMC, Flow Cytometry Research Facility) were used to sort transfected and infected cell lines according to GFP or RFP protein expression. Sorted cells were then collected for plating based on detection of GFP and/or RFP signals.
Virus production
Virus production of pRetroX-Tet-on Advanced, pRetroX-Tight-Pur and pMIG retroviruses was performed in 293GP packaging cells. pCW57.1 lentivirus was produced in 293T packaging cells. Approximately 1 × 106 293T cells were transfected with 2 µg of pCW57.1, 1.5 µg of psPAX2 (Addgene #12260), and 0.5 µg of pMD2.G (Addgene #12259) in a 60 mm plate. Virus was harvested 48 h post transfection. Virus infection in HCT116 cells was performed using10 µg/mL polybrene (Santa Cruz, NC9840454).
Inducible viral gene expression
In order to make inducible Tet-on Advance cell lines OctaKO, Octa/BCL-xl KO, Octa/MCL-1 KO, and Octa/BAX/BAK KO cells were infected with pRetroX-Tet-on Advanced virus in 35 mm plates and plated for single clones in 15 cm plates. Clones were screened for Tet-R expression via western analysis. Single clones expressing Tet-R were used for further experiments. Octa/MCL-1/BAX KO, Octa/MCL-1/BAK KO and Octa/MCL-1/BAX/BAK KO cells were infected with mpMIG-Tet-R virus in 35 mm plates. Infected pools were sorted for GFP-positive cells. Sorted pools were used for further experiments. 1 × 105 cells of Advanced single clones or Tet-R-sorted pools were subsequently infected with pRetroX-Tight-Pur viruses containing the desired gene. Cells were plated and infected in a 35 mm plate using 1:1 ratio of virus and media and 10 µg/mL of polybrene. Infected pools were then selected with 1 µg/mL of puromycin for 24–48 h. Selected pools were used for inducible gene expression assays using 2 µg/mL of Doxycycline.
Genotyping
Genomic DNA was isolated from knockout cell lines using DNeasy Blood and Tissue kit (Qiagen, #69504), and subjected to PCR to amplify genomic target regions of interest using Phusion or Taq polymerase (New England Biolabs, #E0553S and #M0273L) as previously described.23 UNMC High-Throughput DNA Sequencing and Genotyping Core Facility performed Sanger sequencing for genomic PCR products and pGEM plasmids.
Apoptosis assays
Cells (4–5 × 105) were seeded in 35 mm plates 16–20 h prior to apoptosis treatments. Cells were treated with human recombinant TRAIL (25 ng/mL) and/or treated with Doxycycline (2 µg/mL) to induce pro-apoptotic gene expression before being harvested for analysis. For detection of PARP cleavage in treated samples, whole cell lysates were prepared as described above and subjected to western blot with PARP antibody. z-VAD-FMK (40 µM) was used in the indicated assays to allow for protein expression to be detected by immunoblot. Quantification of apoptotic cells according to nuclear morphology was carried out as previously described.44 Cells were stained after apoptotic treatment with 1 μg/mL Hoechst 33342 (Molecular Probes, H-3570) for 30 min at 37 °C. Pictures were taken for three random viewing areas containing 400–800 cells for each plate. Two random zones containing 200–300 cells were selected from each viewing area for cell counting. The percentage of cells undergoing nuclear condensation in each plate was determined by the sum of all zones.
CASPASE 3/7 activity assay
Cells were seeded 48 h prior to treatment in a 96-well plate at a concentration of approximately 1 × 104 cells per well. Thirty microliters of Apo-ONE Caspase 3/7 reagent (Promega, no. G7790) was added to 50 μL of medium in each well after treatment. Plates were shaken in the dark for 1 h to achieve complete cell lysis. Plates were analyzed at 485-nm excitation and 520 emission with a POLARstar Optima microplate reader (BMG Labtech).
BAX dimerization assay
This assay was modified from an assay in a previous study.45 Cells were treated with Doxycycline (2 µg/mL) in the presence of z-VAD-FMK (40 µM) and harvested at the indicated time points. The harvested cells were then re-suspended in lysis buffer (20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1% CHAPS, PH = 7.4) with protease inhibitors (5 mg/mL pepstatin A, 10 mg/mL leupeptin) and 0.1 mM PMSF, and rotated at 4 °C for 1 h. After centrifugation at 22,000 × g for 10 min, the supernatant was collected and split into two halves. BMH (1,6-Bismaleimidohexane, Thermo Fisher Scientific, 22330) was added to the cell lysates in one half to a final concentration of 0.2 mM and mixed. The lysates with or without BMH were incubated at room temperature for 1 h before 4× SDS loading buffer was added to stop the reaction. The samples were then subjected to SDS-PAGE, and analyzed by western blotting with anti-BAX or anti-GFP antibodies.
Supplementary information
Acknowledgements
We are grateful for Keith Johnson and Robert Lewis for helpful discussion and advice. We are also grateful for the UNMC DNA sequencing and flow cytometry facilities for the excellent technical help. This work was supported by a pilot grant from the Nebraska Center for Cellular Signaling (P20-GM103489) and NIH Grants R03CA205496 and R01GM118437 to X.L.
Author contributions
K.H., K.L.O, and X.L. designed the experiments and wrote the manuscript. K.H. and K.L.O. carried out all the experiments. J.L., W.Z., N.H., X.P., W.W., L.S, G.B., Z.L. and L.Z. created some of the cell lines and reagents.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Kai Huang, Katelyn L. O’Neill
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0231-y.
References
- 1.Adams JM, Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25:27–36. doi: 10.1038/cdd.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 3.Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 5.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 6.Kim H, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 7.Czabotar PE, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 12.Kuwana T, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 14.Sarosiek KA, et al. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol. Cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du H, et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J. Biol. Chem. 2011;286:491–501. doi: 10.1074/jbc.M110.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockings C, et al. Bid chimeras indicate that most BH3-only proteins can directly activate Bak and Bax, and show no preference for Bak versus Bax. Cell Death Dis. 2015;6:e1735. doi: 10.1038/cddis.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen HC, et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat. Cell Biol. 2015;17:1270–1281. doi: 10.1038/ncb3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luna-Vargas MPA, Chipuk JE. Physiological and pharmacological control of BAK, BAX, and beyond. Trends Cell Biol. 2016;26:906–917. doi: 10.1016/j.tcb.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai H, et al. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J. Cell Biol. 2011;194:39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis SN, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willis SN, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 22.Edlich F, et al. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Neill KL, Huang K, Zhang J, Chen Y, Luo X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016;30:973–988. doi: 10.1101/gad.276725.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez H, et al. Perturbation of the Bcl-2 network and an induced Noxa/Bcl-xL interaction trigger mitochondrial dysfunction after DNA damage. J. Biol. Chem. 2010;285:15016–15026. doi: 10.1074/jbc.M109.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, et al. Selective involvement of BH3-only proteins and differential targets of Noxa in diverse apoptotic pathways. Cell Death Differ. 2011;18:864–873. doi: 10.1038/cdd.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pang X, et al. The carboxyl-terminal tail of Noxa protein regulates the stability of Noxa and Mcl-1. J. Biol. Chem. 2014;289:17802–17811. doi: 10.1074/jbc.M114.548172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 30.Kotschy A, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 31.Huang K, et al. Cleavage by caspase 8 and mitochondrial membrane association activate the BH3-only protein bid during TRAIL-induced apoptosis. J. Biol. Chem. 2016;291:11843–11851. doi: 10.1074/jbc.M115.711051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs. Trends Biochem. Sci. 2014;39:101–111. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 34.Llambi F, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walensky LD, Gavathiotis E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem. Sci. 2011;36:642–652. doi: 10.1016/j.tibs.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moldoveanu T, et al. BID-induced structural changes in BAK promote apoptosis. Nat. Struct. Mol. Biol. 2013;20:589–597. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borner C, Andrews DW. The apoptotic pore on mitochondria: are we breaking through or still stuck? Cell Death Differ. 2014;21:187–191. doi: 10.1038/cdd.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu YT, Youle RJ. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J. Biol. Chem. 1998;273:10777–10783. doi: 10.1074/jbc.273.17.10777. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 40.Fletcher JI, et al. Apoptosis is triggered when prosurvival Bcl-2 proteins cannot restrain Bax. Proc. Natl Acad. Sci. USA. 2008;105:18081–18087. doi: 10.1073/pnas.0808691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 42.Greaves G, et al. BH3-only proteins are dispensable for apoptosis induced by pharmacological inhibition of both MCL-1 and BCL-XL. Cell Death Differ. 2019;26:1037–1047. doi: 10.1038/s41418-018-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitti Robert M., Marsters Scot A., Ruppert Siegfried, Donahue Christopher J., Moore Alison, Ashkenazi Avi. Induction of Apoptosis by Apo-2 Ligand, a New Member of the Tumor Necrosis Factor Cytokine Family. Journal of Biological Chemistry. 1996;271(22):12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 44.George NM, Evans JJ, Luo X. A three-helix homo-oligomerization domain containing BH3 and BH1 is responsible for the apoptotic activity of Bax. Genes Dev. 2007;21:1937–1948. doi: 10.1101/gad.1553607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang X, et al. Active Bax and Bak are functional holins. Genes Dev. 2011;25:2278–2290. doi: 10.1101/gad.171645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.