Abstract
Deciphering the histone code has illustrated that acetylation or methylation on the same residue can have analogous or opposing roles. However, little is known about the interplay between these post-translational modifications (PTMs) on the same nonhistone residues. We have recently discovered that N-terminal acetyltransferases (NATs) and N-terminal methyltransferases (NRMTs) can have overlapping substrates and identified myosin regulatory light chain 9 (MYL9) as the first confirmed protein to occur in either α-amino-methylated (Nα-methyl) or α-amino-acetylated (Nα-acetyl) states in vivo. Here we aim to determine if these PTMs function similarly or create different MYL9 proteoforms with distinct roles. We use enzymatic assays to directly verify MYL9 is a substrate of both NRMT1 and NatA and generate mutants of MYL9 that are exclusive for Nα-acetylation or Nα-methylation. We then employ eukaryotic cell models to probe the regulatory functions of these Nα-PTMs on MYL9. Our results show that, contrary to prevailing dogma, neither of these modifications regulate the stability of MYL9. Rather, exclusive Nα-acetylation promotes cytoplasmic roles of MYL9, while exclusive Nα-methylation promotes the nuclear role of MYL9 as a transcription factor. The increased cytoplasmic activity of Nα-acetylated MYL9 corresponds with increased phosphorylation at serine 19, a key MYL9 activating PTM. Increased nuclear activity of Nα-methylated MYL9 corresponds with increased DNA binding. Nα-methylation also results in a decrease of interactions between the N-terminus of MYL9 and a host of cytoskeletal proteins. These results confirm that Nα-acetylation and Nα-methylation differentially affect MYL9 function by creating distinct proteoforms with different internal PTM patterns and binding properties.
Introduction
Acetylation and methylation are major post-translational modifications (PTMs) of the histone code and of protein regulation in general [1]. Acetylation most commonly occurs on lysine side chains and replaces the native positive charge with a neutral charge. This charge alteration has been proposed to facilitate protein–protein interactions while disrupting protein–nucleic acid interactions [2,3]. Methylation does not alter the positive charge of lysine or arginine side chains and has been shown to facilitate both protein–DNA and protein–protein interactions [4,5]. Histone acetylation alters transcriptional rates by recruiting bromo and Yeats domain-containing transcription factors to chromatin [6,7], while histone methylation recruits a large number of protein domains to chromatin, including chromo, PHD, Tudor, and PWWP domains [6,8]. Though these reader domains were originally described in the context of histones, some are now emerging as readers of nonhistone proteins as well [9,10].
Based on biological context, histone acetylation or methylation on the same residue can have analogous or opposing functions. Histone H3 lysine 27 (H3K27) can be either methylated or acetylated [11–15]. The trimethylated form is linked to transcriptional repression, and is thought to exert its functional effect by blocking binding of the histone acetyltransferases p300 and CBP, preventing an accumulation of acetylation at enhancers that is required for gene activation [12,14]. H3K27 acetylation has an opposing effect, favoring the activation of enhancers and gene transcription [13]. In contrast, histone H3 lysine 4 (H3K4) can also be methylated or acetylated, but both promote transcriptional activation [15].
Alpha-amino PTMs (Nα-PTMs), occurring on the free protein alpha-amino group rather than on residue side chains, are a widely occurring class of modification whose role in protein regulation remains poorly understood [16–18]. The most prevalent Nα-PTMs, acetylation and methylation, were originally considered to occur on mutually exclusive substrates based on consensus sequence restrictions [16,19,20]. Updated consensus sequences and the identification of both Nα-acetyl and Nα-methyl proteoforms of myosin regulatory light chain 9 (MYL9) have led to the recognition that more than 100 proteins are prospective substrates for such dual modification [21]. Here, we investigate for the first time whether Nα-acetylation and Nα-methylation provide similar or opposing regulation of a common substrate, MYL9, and provide groundwork towards a general understanding of how these Nα-PTMs work coordinately to govern protein function.
MYL9 is a regulatory subunit of the force-generating ATPase nonmuscle myosin II (NMII) [22]. It associates with actin filaments to govern cytoskeletal dynamics and is subsequently involved in cell shape establishment, polarity, adhesion, migration, and signal mechanotransduction [22–24]. The activity of NMII is largely regulated by PTMs of MYL9 [24–26]. Multiple kinases associated with the Rho family of GTPases and the Ca2+ responsive myosin light chain kinase (MLCK) phosphorylate MYL9 at threonine 18 and serine 19 [24,26–28]. Phosphorylation at these residues increases the association of NMII with actin filaments as well as the ATPase activity of the myosin head group [22,25,27]. MYL9 has also been found to play a unique role in the nucleus, as a transcriptional activator of intercellular adhesion molecule 1 (ICAM1) [29]. ICAM1 attracts leukocytes to endothelium and promotes inflammation [30]. MYL9 interacts with ICAM1 promoter DNA directly and with RNAPII and TFIIB, members of the core transcriptional machinery [29]. How the distinct nuclear and cytoplasmic roles of MYL9 are regulated is not yet understood.
Nα-acetylation is catalyzed by the cytoplasmic N-terminal acetyltransferase (NAT) family of enzymes and is thought to occur on up to 80% of the human proteome [17,31]. NatA is the family member with the widest substrate specificity, and its substrate recognition sequence makes MYL9 a prospective target [19]. Diverse, substrate-specific impacts of Nα-acetylation on protein biochemistry and function have been described. It has been shown to promote protein degradation by acting as a docking site for E3 ubiquitin ligases, to be directly involved in protein–protein interactions by facilitating binding in hydrophobic pockets, to assist with proper protein folding and prevention of aggregation, and to increase protein localization to the plasma membrane [31–35].
Nα-methylation is catalyzed by the N-terminal RCC1 methyltransferases 1 and 2 (NRMT1 and NRMT2), the only known eukaryotic alpha-amino methyltransferases [36,37]. These enzymes are found in the nucleus and have over 300 predicted substrate proteins [21,36,37]. While NRMT2 is a monomethylase, NRMT1 is a distributive trimethylase [36,37]. NRMT1 shows more widespread and higher expression across cell types than NRMT2, and trimethylation is the predominant form of Nα-methylation found in cells [36,37]. Until recently, Nα-methylation was thought to be a general promoter of protein stability, as it was originally identified as a modification that protected cytochrome c from degradation [38,39]. Nα-methylation also results in a pH-independent positive charge on the N-terminus and facilitates protein–DNA interactions of substrates such as RCC1, CENP-A, CENP-B, and DDB2 [16,20,40–44].
While we have previously shown that MYL9 in cells can be found in both the Nα-acetylated and Nα-methylated forms [21], we now confirm that MYL9 is a direct substrate of both NatA and NRMT1. We utilize the consensus sequence requirements of these enzymes to generate mutants of MYL9 that select for exclusive Nα-acetylation or Nα-methylation. We use a photoswitchable fluorescent tag to demonstrate that despite the prevailing dogma in the field, Nα-acetylation and Nα-methylation do not alter the half-life of MYL9. Rather, we find that Nα-acetylation promotes the cytoplasmic roles of MYL9 and its phosphorylation at serine 19. Nα-methylation promotes the nuclear role of MYL9 and its DNA binding. Neither Nα-acetylation nor Nα-methylation is recognized by readers of histone acetylation or methylation. Surprisingly, Nα-methylation actually perturbs MYL9 protein–protein interactions, including those with many cytoskeletal proteins. Taken together, these data show for the first time that Nα-acetyl-MYL9 and Nα-methyl-MYL9 are different proteoforms with different cellular functions, and that in this context acetylation is permissive for protein–protein interactions and internal phosphorylation, while methylation favors interaction with DNA.
Materials and methods
Recombinant protein expression and in vitro enzyme assays
Human NRMT1 (rhNRMT1) and NAA10 (rhNAA10) were expressed as His6-tagged recombinant proteins and purified as previously described [45,46]. Briefly, protein ORFs (GE Dharmacon, Marlborough, MA) were amplified and subcloned into the pET15b vector (EMD Millipore, Billerica, MA), expressed in BL21 Star (DE3) Escherichia coli (Thermo Scientific, Waltham, MA), and purified on Ni2+-NTA beads (Qiagen, Hilden, Germany).
In vitro methyltransferase experiments were carried out using the MTase-Glo Methyltransferase Assay (Promega, Madison, WI) following manufacturer guidelines. Briefly, 0.2 μM rhNRMT1 was incubated at room temperature (RT) with synthetic peptide substrates corresponding to the 14 N-terminal amino acids of wild type (WT) or mutant MYL9 (Figure 1A) (AnaSpec, Fremont, CA) in the presence of 40 μM s-adenosyl methionine (SAM). Peptide substrates were used at concentrations ranging from 0 to 160 μM. Reactions were stopped at 20 min by addition of trifluoroacetic acid. Methyltransferase activity was then measured by the accumulation of a luminescent signal, which was generated by reaction of a detection reagent with the methyltransferase reaction product s-adenosyl-L-homocysteine (SAH).
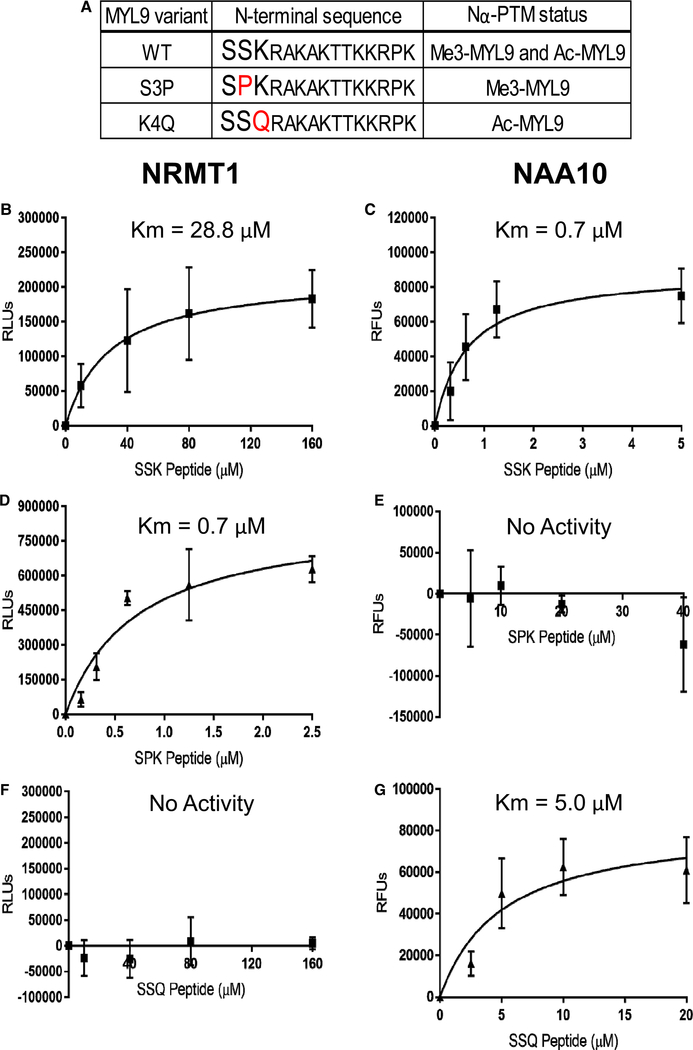
Figure 1. N-terminal mutants of MYL9 select for Nα-methylation or Nα-acetylation.
(A) WT MYL9 (SSK) has been found in Nα-methylated and Nα-acetylated forms. The S3P (SPK) MYL9 mutant is Nα-methylated, and the K4Q (SSQ) MYL9 mutant is Nα-acetylated. In vivo, the initiator methionine of WT MYL9 is cleaved to reveal the N-terminal sequence shown. (B–G) in vitro methyltransferase and acetyltransferase assays were performed to determine the KM of NRMT1 or NAA10 (the catalytic subunit of NatA) when using peptides corresponding to the 14 N-terminal amino acids (after Met cleavage) of WT and mutant MYL9 as substrates. (B) The KM of NRMT1 with WT (SSK) MYL9 was determined to be 28.8 μM. (C) NAA10 with WT (SSK) MYL9 had a KM of 0.7 μM. (D) SPK MYL9 was confirmed to be a preferred substrate of NRMT1 with a KM of 0.7 μM. (E) NAA10 showed no activity with SPK MYL9 up to 40 μM. (F) SSQ MYL9 Figure 1. N-terminal mutants of MYL9 select for Nα-methylation or Nα-acetylation. Part 2 of 2 was not a substrate of NRMT1, showing no activity up to 160 μM. (G) The KM of NAA10 with SSQ MYL9 was 5.0 μM. n = 3 for all experiments. All error bars represent standard deviation.
In vitro acetyltransferase experiments were performed using the Enzo Acetyltransferase Activity Kit (Enzo Life Sciences, Farmingdale, NY) following manufacturer guidelines. Briefly, 0.1 μM rhNAA10 was incubated with N-terminal peptides of WT or mutant MYL9 at RT in the presence of excess acetyl-CoA. The reaction was stopped after 30 min by addition of ice-cold isopropyl alcohol. Acetyltransferase activity was measured by reading fluorescence (Ex 380 nm/Em 520 nm) generated by the reaction of a detection reagent with the acetyltransferase reaction product CoA.
All readings were taken on a Cytation5 cell imaging multi-mode reader (BioTek, Winooski, VT). For both methyltransferase and acetyltransferase assays, background signal was measured by the inclusion of no substrate control reactions and was subtracted from experimental reactions. Background subtracted signal intensities at varying substrate concentrations were then fit to a model of Michaelis–Menten enzyme kinetics and a KM determination was made using GraphPad Prism 7 software (San Diego, CA).
Molecular cloning, mutagenesis, and lentivirus production
To generate the C-terminally tagged MYL9-Dendra2 fusion protein, the MYL9 protein ORF (GE Dharmacon) was amplified and subcloned into the pDendra2-N vector (Clontech, Mountain View, CA) using XhoI and HindIII restriction sites. Site-directed mutagenesis was performed using the Quikchange system (Agilent Technologies, Santa Clara, CA) to generate S3P (SPK) and K4Q (SSQ) mutant MYL9-Dendra2 fusion proteins. WT, SPK, and SSQ MYL9-Dendra2, as well as Dendra2 alone, were then amplified and subcloned into the pCDH-EF1-MCS-IRES-Puro lentiviral expression vector (System Biosciences, Palo Alto, CA) using XbaI and NheI restriction sites. The following forward primers and their reverse compliments were used for mutagenesis: S3P: 5′-GATCTCGAGATGTCCCCCAAGCGGGCCAAAGC-3′ and K4Q: 5′-CGAGATGTCCAGCCAGCGGGCCAAAGCC-3′. To generate C-terminally tagged MYL9-FLAG proteins, WT, SPK, and SSQ MYL9 were amplified from the MYL9-pDendra2 constructs and subcloned into the pWPI lentiviral expression vector (Addgene, Cambridge, MA) using 5′ and 3′ PmeI restriction sites. The following 3′ primer included the FLAG nucleotide sequence followed by a stop codon: 5′-CGGTTTAAACTCATTTATCATCATCATCTTTATAATCGTCGTCTTT ATCCTTGGCGC-3′ Lentivirus was generated through co-transfecting HEK293T cells with 50 μg of expression plasmid (pCDH or pWPI from above), 37.5 μg psPAX2 packaging vector, and 15 μg pMD2.G envelope plasmid using calcium phosphate transfection as previously described [47]. Viral supernatants were collected forty-eight hours after transfection, concentrated with 100 KDa molecular mass cut-off filters (Millipore Sigma, Burlington, MA), and titered in HEK293T cells. GFP expressed by the pWPI vector and Dendra2 expressed by the pCDH vector was used for titering.
Cell culture, transgene expression, and small molecule treatment
HEK293T human embryonic kidney and NIH 3T3 mouse embryonic fibroblast cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Life Technologies, Grand Island, NY) with 10% fetal bovine serum (FBS; Atlanta Biologicals, Atlanta, GA) and 1% penicillin–streptomycin (P/S; Life Technologies). HCT116 human colorectal carcinoma and CRISPR/Cas9-mediated NRMT1 knockout (KO) HCT116 cell lines were cultured in McCoy’s 5A Modified Medium (Life Technologies) supplemented with 10% FBS and 1% P/S. NRMT1 KO cells were previously generated and verified by sequencing and protein expression [45]. Maintenance of NRMT1 loss was verified by Western Blot (Figure 4E). All cells were grown on tissue culture treated plastic and maintained at 37°C and 5% CO2. HEK293T, NIH 3T3, and HCT116 cell lines were a generous gift from Dr. Ian Macara, Vanderbilt University.
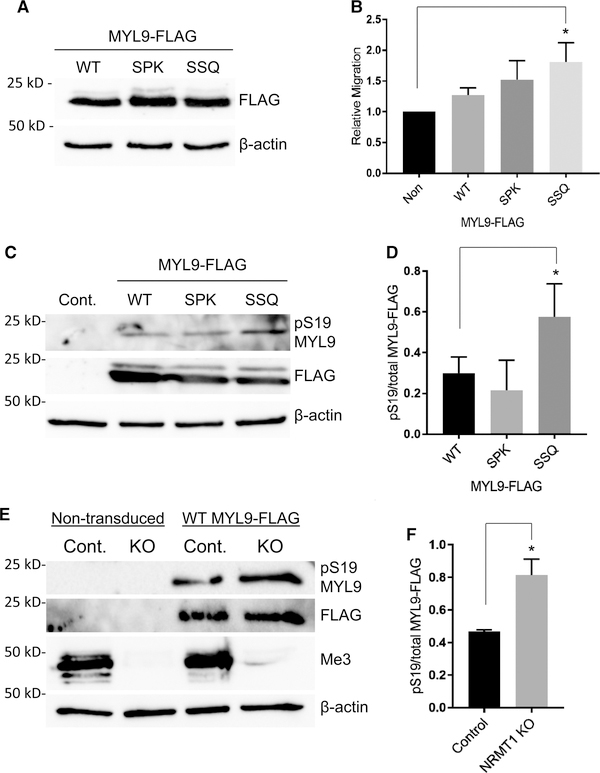
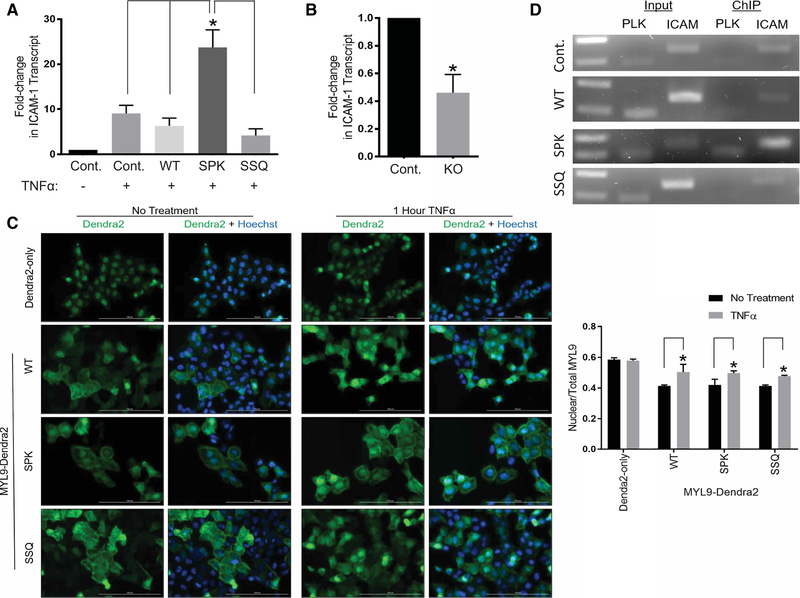
Figure 4. Nα-acetylation promotes cell migration and serine 19 phosphorylation.
(A,B) HCT116 cells were transduced with WT or mutant MYL9-FLAG and transwell migration assays were performed. (A) Western Blotting confirms that MYL9 variants were expressed at equal levels. β-actin was used as a loading control. (B) Only cells expressing the nonmethylatable SSQ MYL9 mutant showed significantly greater migration than nontransduced control cells (P = 0.01), indicating enhanced cytoskeletal activity (Control — 1.00, WT — 1.27 ± 0.10, SPK — 1.52 ± 0.25, SSQ — 1.81 ± 0.26). (C) Representative blot showing increased pS19 of the nonmethylatable SSQ MYL9 mutant after calcimycin treatment. FLAG blot used to determine total MYL9 protein levels. (D) SSQ MYL9 had a significantly greater ratio of pS19 to total protein than SPK MYL9 (P < 0.05; WT — 0.30 ± 0.06, SPK — 0.21 ± 0.12, SSQ — 0.58 ± 0.13). (E) Representative blot showing increased pS19 of WT MYL9 in cells that lack Nα-methylation (Me3). (F) WT MYL9 in NRMT1 KO cells had a significantly greater ratio of pS19 to total protein than WT MYL9 in control cells (P < 0.005; Control — 0.47 ± 0.01, NRMT1 KO — 0.81 ± 0.08). One-way ANOVA with Tukey’s multiple comparisons test was used for analysis of results in B and D. Results in F were analyzed by a Student’s two-tailed t-test. n = 3 for all experiments. All error bars represent standard deviation.
For experiments involving transgene expression, cells were harvested with Trypsin-EDTA (Life Technologies), plated in fresh media, and transduced with lentivirus at a multiplicity of infection (MOI) of 2. Transgene expression was confirmed and cells were used for experiments 3–4 days after transduction. Cells transduced with the pCDH vector were selected for by treatment with 2 μg/ml puromycin 48 h after transduction, and were used for experiments 48 h after treatment. For experiments involving small molecule treatment, cells were cultured to 80% confluence, and media was changed to fresh media containing the desired small compound. Calcium ionophore A23187 (calcimycin; Sigma–Aldrich, St. Louis, MO) was resuspended in DMSO and used at a final concentration of 0.1 μM. Cells were treated with calcimycin for 45 min to induce phosphorylation of MYL9 by MLCK. Recombinant human tumor necrosis factor alpha (TNFα; Life Technologies) was resuspended in sterile, distilled water and used at a final concentration of 20 ng/ml. Cells used in transcriptional studies were treated for 2 h, while cells used for localization and promoter binding studies were treated for 1 h.
Half-life measurement and modeling
25K HEK293 cells expressing Dendra2 or Dendra2-MYL9 fusion proteins were plated in triplicate in blackwalled, clear-bottom 96-well tissue culture plates and given 24 h to adhere. Experiments were then carried out in a Cytation 5 cell imaging multi-mode reader maintained at 5% CO2 and 37°C. Cells were imaged prephotoconversion at 20× in the GFP and RFP channels, followed by photoconversion through exposure to the DAPI laser line (UV) at maximum intensity for 16 sec. Following photoconversion, cells were imaged in the RFP channel every 4 h for 48 h, imaging settings (LED intensity, integration time, and gain) were maintained throughout experiments. Sum RFP fluorescence intensity values were calculated from post-photoconversion images at each time point, and then were plotted relative to the 0 h intensity. Three to four experiments were performed for each group, and MYL9-Dendra decay curves were then generated in GraphPad Prism using a one-phase exponential decay model. Initial y values were set to 1 for all curves and curve plateaus (y-value at infinite time) were set to 0. Decay curves for each MYL9-Dendra variant were compared with WT in a pairwise fashion to determine if the data sets could be adequately fit by a single curve or if independent curves were required. An extra sum-of-squares F test was used to compare the goodness of fit of shared and independent curves, and subsequently determine the probability with which the shared curve represents both data sets. MYL9 half-life was calculated as ln(2)/k, with k being the rate constant (h−1) generated by the exponential decay model.
For cycloheximide experiments, 1 × 106 HCT116 cells or HCT116 cells transduced with lentivirus expressing WT-MYL9-FLAG, SPK-MYL9-FLAG, or SSQ-MYL9-FLAG were plated in 6-well tissue culture plates 24 h prior to cycloheximide treatment. 100 μg/ml cycloheximide (Millipore Sigma) was added, and cells were harvested in 100 μl cell lysis buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 1% NP-40, 7 mM BME, 1 mM PMSF, 25 μg aprotinin, and 25 μg leupeptin) at indicated time points. Protein was quantified with Pierce 660 nM Protein Assay (Thermo Scientific) and 20 μg total protein was loaded per lane. Samples were analyzed by Western blots as described below.
Cell spreading
Two-well chamber slides (Thermo Scientific) were coated with 3 μg/cm2 fibronectin (Corning, Corning, NY) in Tris-buffered saline for 1 h at RT. The remaining solution was removed and slides were blocked with 5 mg/ml bovine serum albumin (BSA; Research Products International, Mt. Prospect, IL) in phosphate-buffered saline. Control and NIH 3T3 cells expressing WT, SPK, or SSQ MYL9-FLAG were grown to confluence, trypsinized, counted, and resuspended at 100K cells/ml in fresh media. Cells were then placed on ice for 20 min to promote rounding. 70K cells were then plated per chamber, and chamber slides were immediately placed in a Cytation 5 cell imaging multi-mode reader with CO2 maintained at 5% in the sample chamber. The sample chamber was raised from RT to 37°C, with time 0 being defined as when 37°C was reached. Cells were then imaged with a 20× phase contrast objective every 15 min for 1 h. The surface area was quantified for the four largest cells per image at each time point by tracing cell outlines using the ImageJ 1.47v (NIH, Bethesda, MD) plug-in NeuronJ version 1.4.3 [48]. Twelve cell areas taken from three independent experiments were used to generate the mean cell surface area for each sample at each time point.
Transwell migration
Cell migration was measured using a 96-well basement membrane extract (BME) cell invasion assay (Trevigen, Gaithersburg, MD) as previously described [40]. Briefly, control and WT, SPK, or SSQ MYL9-FLAG expressing HCT116 cells were serum-starved for 24 h in McCoy’s 5A Modified Medium with 0.1% BSA. 50K cells in serum-free media were then plated on 0.25× BME in triplicate in the upper chamber of a transwell plate. Media supplemented with 1% FBS was plated in the lower chamber of transwells to serve as a chemoattractant. Migration was allowed to proceed for 24 h, nonmigrating cells were washed away, and migrating cells were stained using Calcein-AM. Fluorescence at 485 nm excitation, 520 nm emission was used to quantify the degree of cell migration.
Western blots
Twenty microgram of total protein per sample was separated on 10% SDS–PAGE gels and transferred to nitrocellulose membranes using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad, Hercules, CA). Nitrocellulose membranes were blocked for one hour in 5% w/v nonfat dry milk in TBS + 0.1% Tween 20 (TBST) at RT. Primary and secondary antibodies were also diluted in the 5% dry milk solution. Dilutions used for primary antibodies were: rabbit anti-FLAG (1:500; Invitrogen, Carlsbad, CA), HRP-conjugated mouse anti-FLAG (1:1000; Sigma–Aldrich), rabbit anti-β-actin (1:1000; Cell Signaling Technologies, Danvers, MA), rabbit anti-pS19 MYL9 (1:1000; Cell Signaling Technologies), rabbit anti-Nα-trimethyl RCC1 (1:10,000) [20], rabbit anti-PSMD7 (1:1000; Origene, Rockville, MD), rabbit anti-cofilin (1:1000; abcam, Cambridge, MA), and rabbit anti-HEXIM2 (1:1000; abcam). Membranes were rocked in primary antibody overnight at 4°C and washed three times for 15 min in TBST. Membranes were rocked in secondary antibody for 1 h at RT and washed three times for 15 min in TBST. The secondary antibody used was donkey anti-rabbit (1:5000; Jackson ImmunoResearch, West Grove, PA). Blots were developed using KPL LumiGLO Peroxidase Chemiluminescent Substrate (VWR, Radnor, PA) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) and a ChemiDoc imaging system (Bio-Rad). Densitometry was performed using ImageJ 1.47v software (NIH).
Real time PCR analysis
Cells were lysed in TRIzol (Life Technologies) and RNA was then extracted in chloroform, pelleted in isopropanol, and washed with ethanol. The final pellet was air dried and resuspended in 100 μl sterile water. RNA concentration was determined with the Cytation 5 plate reader (BioTek). One microgram of RNA was taken through the SuperScript First-Strand Synthesis System (Life Technologies) to generate cDNA according to manufacturer guidelines. Two microgram of cDNA from each sample was used as template with SYBR Green PCR Master Mix (Bio-Rad) in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Transcript expression levels were compared using the ΔΔCT quantification method, with GAPDH serving as a control. Melt curves were performed to confirm the presence of single reaction products. Primers (Integrated DNA Technologies, Coralville, IA) used were:
ICAM1 forward 5′-GTAGCAGCCGCAGTCATAAT-3′
ICAM1 reverse 5′-GGGCCTGTTGTAGTCTGTATTT-3′ GAPDH forward 5′-ACAGCCTCAAGATCATCAGCAA-3′ GAPDH reverse 5′-CCATCACGCCACAGTTTCC-3′.
Immunofluorescence
Dendra2 or MYL9-Dendra2 expressing HCT116 cells were analyzed for MYL9 cellular compartmentalization either before or 1 h post-TNFα treatment. At RT, cells were fixed with 4% paraformaldehyde for 10 min, quenched in 50 mM ammonium chloride for 10 min, permeabilized with 0.3% Triton X-100 for 5 min, and blocked with 3% BSA for 1 h. Mouse anti-Dendra2 primary antibody (Origene) was diluted 1:150 in 1% BSA and samples were incubated overnight at 4°C. Samples were washed three times in 1 ml of PBS. Alexa Fluor 488 conjugated goat anti-mouse secondary antibody (Thermo Scientific) was diluted 1:2000 in 1% BSA and added to the samples for 1 h at RT. Cells were washed three times in 1 ml of PBS and counterstained with Hoechst dye (AnaSpec) diluted 1:50,000 in PBS. Cells were washed three times in 1 ml of PBS and slides were mounted with Fluoromount-G (Southern Biotech, Birmingham, AL). Imaging was performed on a Cytation 5 cell imaging multi-mode reader with a 40× objective and image analysis was performed using Gen5 software (BioTek). Total Dendra2 signal was quantified by the sum green fluorescence intensity of images. To quantify nuclear Dendra2, the sum green fluorescence intensity overlapping with Hoechst signal was quantified.
Chromatin immunoprecipitation
Control and WT, SPK, or SSQ MYL9-FLAG expressing HCT116 cells were treated with TNFα for 1 h. 1 × 107 cells were cross-linked with 1% formaldehyde for 10 min at RT, quenched with 1× glycine for 5 min at RT, resuspended in 600 μl ChIP lysis buffer [150 mM NaCl, 25 mM Tris–HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate (NaDoc)], and sonicated 6 × 10 s at 30% amplitude to fragment DNA. 10% of total sheared lysate was saved as an input sample. MYL9-FLAG was then immunoprecipitated from lysates overnight at 4°C using 20 μl M2 agarose anti-FLAG beads (Millipore Sigma) that were pre-blocked in herring sperm DNA (Thermo Scientific). After immunoprecipitation, beads were washed one time in High salt ChIP wash buffer (500 mM NaCl, 20 mM Tris–HCl, pH 8.0, 0.1% SDS, 1% NP-40, 2 mM EDTA), one time LiCl ChIP wash buffer (0.25 M LiCl, 10 mM Tris–HCl, pH 8.0, 1% NaDoc, 1% NP-40, 1 mM EDTA) and two times Tris–EDTA (TE). Bound material was eluted 2× in 100 μl elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at RT. De-cross-linking was performed by adding 8 μl 5M NaCl to each sample and incubating overnight at 65°C. Eluates were then treated with 1 μl of RNase A (24 mg/ml stock, Thermo Scientific) and 1 μl of proteinase K (10 mg/ml stock, Millipore Sigma) and incubated at 45°C for 1 h. The remaining immunoprecipitated DNA was isolated using a Qiagen PCR purification kit (Qiagen) and resuspended in 30 μl of TE buffer. Input samples were processed alongside ChIP samples after elution. Equal amounts of purified DNA were amplified by PCR and analyzed by running on 2% agarose gels. PLK-1 served as a control for nonspecific DNA binding. Primers for the PLK-1 (Integrated DNA Technologies) and ICAM1 (Invitrogen) promoters were as follows: ICAM1 forward 5′-CGCCCGATTGCTTTAGCTTG-3′, ICAM1 reverse 5′-GGCTGAGGTTGCAACTCTGA-3′, PLK-1 forward 5′-GGTTTGGTTTCCCAGGCTAT-3′, and PLK-1 reverse 5′-GCTGGGAACGTTACAAAAGC-3′.
Statistical analyses
Statistics were performed with GraphPad Prism 7 software. For experiments involving the comparison of only two groups, a two-tailed Student’s t-test was performed. For experiments involving the comparison of three or more groups, one-way ANOVA was performed with Tukey’s test for multiple comparisons between all groups. For experiments involving comparison of three or more groups at multiple time points, two-way random measures ANOVA with Tukey’s multiple comparisons test between all groups was performed. For experiments involving comparison of three or more groups under two conditions, regular two-way ANOVA with Bonferroni’s multiple comparisons test between all groups was performed. All error bars shown represent standard deviation.
Peptide arrays
Experiments were carried out at the MD Anderson Protein Array and Analysis Core under the direction of Dr. Mark Bedford as previously described [49]. Briefly, arrays of recombinant methyl or acetyl reader domains were immobilized on chips using a high-density arrayer. The methyl reader array consisted of 104 unique domain constructs and the acetyl reader array consisted of 30 unique domain constructs. Biotinylated N-terminal peptides of MYL9 were synthesized (Anaspec) and flowed over the reader chips. Unmodified, mono, and tri methylated MYL9 peptides were tested for binding on the methyl reader array. Unmodified and acetylated MYL9 peptides were tested for binding on the acetyl reader array. Peptides that bound reader domains were then detected using streptavidin–AlexaFluor488 and visualized using a Molecular Probes GenePix 4400A MicroArray Scanner.
SILAC LC–MS/MS
The SILAC LC–MS/MS screen was adapted from a previously described protocol to identify readers of trimethyl-lysine [50]. Briefly, HEK293 cells were grown in normal media (light) or media supplemented with 13C labeled lysine (heavy) for eight passages. This resulted in 98.9% incorporation of isotopically labeled lysine into proteins of cells grown in heavy media as determined by LC–MS/MS. For pulldowns, synthetic 14 amino acid N-terminal peptides of MYL9 were synthesized in an unmodified, Nα-acetylated, Nα-monomethylated, and Nα-trimethylated state (Anaspec). All peptides were synthesized with a C-terminal biotin moiety, allowing them to be linked to M280 streptavidin Dynabeads. Pulldowns with unmodified MYL9 peptide were performed using protein lysate harvested from cells grown in light media. Separate pulldowns were performed with acetylated, monomethylated, or trimethylated MYL9 peptides using heavy isotope labeled protein lysate. Each pulldown was performed with 20 μg of peptide and 300 μg of protein lysate. After pulldowns, beads were washed and bound protein was eluted in Laemmli buffer. For analysis, elutions from each modified peptide pulldown were mixed with an elution from an unmodified peptide pulldown. This generated three samples that each contained light proteins that interacted with unmodified MYL9 and heavy proteins that interacted with one of the forms of modified MYL9, allowing for a quantitative comparison of interactors based on modification state. Samples were resolved by SDS–PAGE, in-gel trypsin digested, and subjected to LC–MS/MS using a NanoAcquity HPLC system interfaced to a ThermoFisher Q Exactive mass spectrometer. LC–MS/MS and subsequent data analysis were performed by MS Bioworks (Ann Arbor, MI). Only proteins that had a SILAC ratio (peptides identified in both light and heavy states) were included for further analysis, resulting in ~650 proteins per sample pair. Interactions of interest were further selected by setting a cut-off of a three-fold change in the SILAC ratio (0.33 > heavy/light > 3). For validation of SILAC results, pulldowns were performed as above using only normal media. After being separated by SDS–PAGE, proteins were transferred to nitrocellulose and taken through Western blot analysis with anti-PSMD7, anti-HEXIM2, or anti-cofilin antibodies.
Results
N-terminal mutants of MYL9 select for Nα-methylation or Nα-acetylation
MYL9 has previously been immunoprecipitated from cells or tissue in both Nα-methyl and Nα-acetyl modification states [21]. The N-terminal methionine of MYL9 is cleaved, revealing a Ser-Ser-Lys (SSK) amino acid sequence that conforms to the consensus sequences of both NRMT1 and NatA (Figure 1A) [19,21]. To confirm these enzymes catalyze the Nα-PTMs of MYL9, in vitro enzymatic assays were performed with rhNRMT1 and rhNAA10, the catalytic subunit of NatA. A peptide substrate corresponding to amino acids 2–15 of MYL9 was used at varying concentrations to generate a KM determination for each enzyme. Both NRMT1 and NAA10 were found to catalyze the modification of MYL9. NRMT1 showed moderate activity with a KM of 28.8 μM (Figure 1B), while NAA10 showed high activity with a KM of 0.7 μM (Figure 1C).
To test if we could select for exclusive Nα-methylation or Nα-acetylation of MYL9, we designed N-terminal mutants of MYL9 based on the consensus sequence requirements for NRMT1 and NatA. The N-terminal amino acid sequence SPK is known to be a favorable substrate for NRMT1, while a proline in the second position of proteins is well described as blocking NatA activity [19,21,37]. Similarly, NRMT1 activity is restricted by a glutamine in the third position, while this amino acid is permissive for NatA activity [19–21]. Based on these data, we utilized 14 amino acid peptides of MYL9 containing either an S3P (SPK MYL9) or K4Q (SSQ MYL9) mutation as substrates in the in vitro enzymatic assays, to see if they could select for the activity of either NRMT1 or NatA (Figure 1A).
Matching the published consensus sequence requirements, NRMT1 showed increased affinity for SPK MYL9 with a KM of 0.7 μM (Figure 1D), while NAA10 showed no activity with SPK MYL9 out to 40 μM of substrate (Figure 1E). SSQ MYL9 showed opposing results, with NRMT1 having no activity on this substrate out to 160 μM of substrate (Figure 1F), and NAA10 maintaining activity with a KM of 5.0 μM (Figure 1G). Taken together, these results demonstrate that the N-terminal consensus sequence of MYL9 is permissive for modification by both NRMT1 and NatA, and that SPK and SSQ mutants of MYL9 select for either Nα-methylation or Nα-acetylation, respectively.
Nα-PTMs do not alter the half-life of MYL9
A major focus of the Nα-PTM field has been on how these modifications affect the stability of proteins. Early studies suggested that Nα-methylation protects proteins from degradation, as this modification was shown to increase the stability of cytochrome c and block degradation by aminopeptidases [38,39]. Nα-acetylation is known to decrease the stability of some proteins by acting as a docking site for E3 ubiquitin ligases, and thus promoting proteolytic degradation [33,51]. However, accumulating evidence suggests that these effects on stability are substrate specific, rather than intrinsic properties of Nα-acetylation and Nα-methylation [18,31].
To see if Nα-PTMs regulate the stability of MYL9, the photoswitchable fluorescent protein Dendra2 was C-terminally tagged to full-length WT, SPK, and SSQ MYL9. Dendra2 in its basal state fluoresces in the GFP channel, but after photoconversion fluoresces in the RFP channel [52,53]. This allows for tracking of half-life by following the rate of red fluorescence decay after photoconversion, and alleviates the need to treat with cytotoxic agents such as cycloheximide [54]. When Dendra2 was expressed alone and photoconverted, the sum red fluorescence intensity was found to be stable over 48 h, verifying the Dendra2 signal does not dissipate over this time frame (Figure 2A,B). When WT and mutant MYL9 were tagged with Dendra2, decay in red fluorescence related to the degradation of MYL9-Dendra2 was observable within 24 h of photoconversion for all variants (Figure 2C). This was confirmed by cycloheximide experiments that also showed observable protein turnover for all MYL9-FLAG variants within 24 h (Supplementary Figure S1).
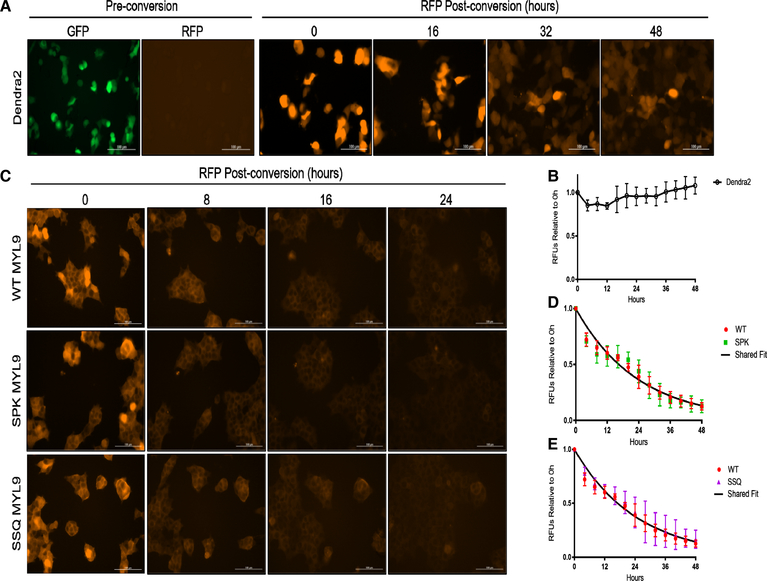
Figure 2. Nα-PTMs do not alter the half-life of MYL9.
(A) Fluorescence images of cells expressing Dendra2 alone. (B) The sum fluorescence intensity of Dendra2 only expressing cells is plotted over 48 h. Individual cells show decreased RFP intensity as cell division occurs, but the sum intensity over the field of view remains constant. (C) Fluorescent images of cells expressing WT or mutant MYL9-Dendra2 out to 24 h. (D,E) Fluorescent decay for WT, SPK, and SSQ MYL9 was plotted and fit to a model of one-phase exponential decay. SPK and SSQ MYL9 were each fit to an individual model of decay and a model of decay shared with WT MYL9. Individual and shared fit models were then evaluated by an extra sum-of-squares F test. For both SPK and SSQ MYL9 it was determined that a shared model of decay with WT MYL9 was as effective as an individual model, indicating no effect on stability. The half-life of MYL9 was determined to be 16.4 h. Scale bars are 100 μM. n = 3–4 for all experiments. All error bars represent standard deviation.
The half-life of WT MYL9-Dendra2 was determined to be 16.4 h (95% confidence interval, 15.49–17.52 h) by fitting fluorescence decay data gathered over 48 h to a model of one-phase exponential decay. To determine if alterations in Nα-modification state could be statistically linked to changes in the rate of MYL9 decay, SPK and SSQ MYL9 fluorescence data were each fit to an individual model of decay as well as a model shared with WT MYL9. These models were compared for best fit, and in each case it was found that a model of decay shared with WT MYL9 was adequate to describe the decay of SPK and SSQ MYL9 (Figure 2D,E). This demonstrates that regulation of MYL9 stability is not a role for Nα-methylation or Nα-acetylation, and helps overturn the perception that the primary role of these modifications is as modifiers of protein half-life.
Nα-acetylation of MYL9 promotes cell spreading
We next turned our attention to how Nα-PTMs regulate the function of MYL9 in the cytoplasm. The primary role of MYL9 in the cytoplasm is regulation of actin cytoskeletal dynamics through modulation of NMII activity [22,24]. We analyzed cell spreading of NIH 3T3 mouse fibroblasts on fibronectin, as fibroblast adhesion to fibronectin is a well-studied process requiring active cytoskeletal rearrangement [55–57]. We hypothesized that Nα-acetylation would facilitate the cytoplasmic role of MYL9 as a cytoskeletal regulator. As such, we expected the Nα-acetylation exclusive SSQ mutant of MYL9 to show the most robust cell spreading.
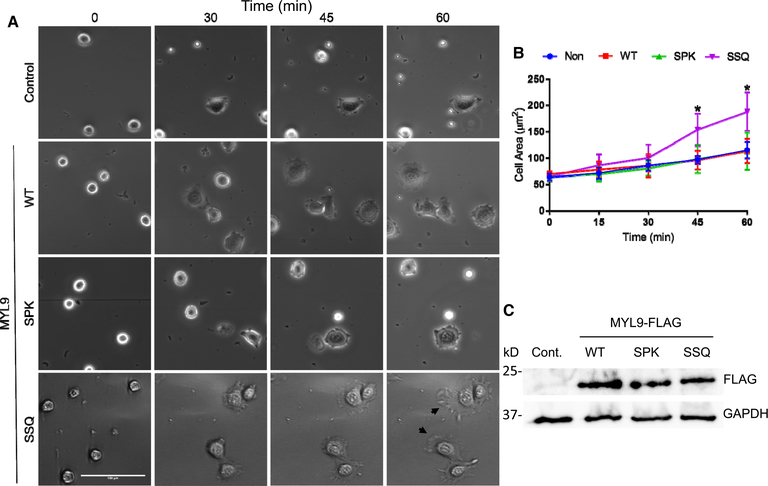
Control NIH 3T3 cells or cells transduced with WT, SPK, or SSQ MYL9-FLAG were plated on 3 μg/ml fibronectin-coated chamber slides and imaged every 15 min for 1 h. Cells were placed on ice prior to plating to promote a uniform distribution of rounded, single cells (Figure 3A, first column). Untransduced, WT MYL9-expressing, and SPK MYL9-expressing cells were just beginning to spread after 1 h and covered very little surface area (Figure 3A,B). However, the Nα-acetylation exclusive SSQ MYL9-expressing cells covered a significantly greater surface area after 1 h (Figure 3B), and only they had readily observable lamellipodia and filopodia that stretched over the fibronectin-coated surface (Figure 3A, black arrows). In order to ensure that expression level differences were not driving our results, we confirmed equal expression of WT, SPK, and SSQ MYL9-FLAG by Western blotting (Figure 3C). As such, it appears that exclusive Nα-acetylation favors the participation of MYL9 in cytoskeletal rearrangement.
Figure 3. Nα-acetylation of MYL9 promotes cell spreading.
(A) NIH 3T3 cells expressing the Nα-methylation deficient SSQ mutant of MYL9 showed considerable cell spreading, with lamellipodia and filopodia (black arrows) readily observable. Scale bar is 100 μM. (B) Cells expressing SSQ MYL9 covered significantly greater area than all other cell lines at 45 (P < 0.01) and 60 (P < 0.001) minutes as determined by two-way random measures ANOVA with Tukey’s multiple comparisons test (all measurements μm2; 45 min: Control — 96.7 ± 6.1, WT MYL9 — 96.4 ± 14.6, SPK MYL9 — 98.4 ± 22.1, SSQ MYL9 — 154.1 ± 25.2; 60 min: Control — 115.1 ± 12.6, WT MYL9 — 113.6 ± 18.7, SPK MYL9 — 113.0 ± 28.9, SSQ MYL9 — 188.3 ± 30.0) (n = 3). (C) Equal expression of MYL9 variants was confirmed through Western Blotting. FLAG was used as a measure of MYL9-FLAG expression. GAPDH was used as a loading control. All error bars represent standard deviation.
Nα-acetylation promotes cell migration and serine 19 phosphorylation
We next sought to confirm the impact of Nα-methylation on the cytoskeletal activity of MYL9 in HCT116 human colon carcinoma cells. These cells have a migratory phenotype, and the nuclear role of MYL9 in ICAM1 transcription has been reported in colon cells, making this cell line suited for the study of both cytoplasmic and nuclear functions of MYL9 [29,58]. To test the role of Nα-modifications of MYL9 in regulating cytoskeletal activity, we measured the migratory potential of nontransduced control cells and cells transduced with WT, SPK, or SSQ MYL9-FLAG. A Boyden chamber design was employed, in which cells were serumstarved, plated on an extracellular matrix extract, and migration towards serum containing media was quantified after 24 h. MYL9 variants were transduced into cells at equal levels so that protein abundance differences would not affect results (Figure 4A). Only HCT116 cells expressing the Nα-methylation deficient SSQ MYL9 mutant showed a significant increase in migration as compared with nontransduced cells (Figure 4B). This suggests that exclusive Nα-acetylation of MYL9 leads to increased cytoskeletal dynamics, as required for migration. This result is consistent with our findings in NIH 3T3 cells, and supports a model in which Nα-acetylation of MYL9 participates in the activation of NMII.
Multiple signaling pathways converge on phosphorylation of MYL9 at serine 19 (pS19) in order to increase NMII activity [22,24]. Because of the requirement for pS19 MYL9 in NMII related activities, including migration and cell spreading, we next tested if the activated cytoskeletal function of SSQ MYL9 is associated with increased levels of pS19. To accomplish this, we treated HCT116 control and MYL9-FLAG (WT and mutant) expressing cell lines with calcimycin, an MLCK activator, and evaluated pS19 levels. We found that after treatment, the nonmethylatable SSQ MYL9 mutant has increased pS19 as compared with WT and SPK MYL9 (Figure 4C). The ratio of pS19 to total MYL9-FLAG was quantified, and it confirmed that SSQ MYL9 shows enrichment of this activating PTM (Figure 4D). This result provides a biochemical basis for the cytoskeletal phenotypes seen in SSQ MYL9-expressing cells and suggests Nα-acetylation is more permissive for S19 phosphorylation than Nα-methylation.
As blocking of phosphorylation by neighboring methylation is a common phenomenon [59,60], we wanted to determine if in addition to Nα-acetylation being permissive to pS19, Nα-methylation could be inhibitory. We thereby assayed the phosphorylation of WT MYL9 in NRMT1 KO HCT116 cells [45]. These cells are deficient in Nα-methylation (Figure 4E), and if Nα-methylation is inhibitory, they should exhibit higher S19 phosphorylation of WT MYL9 than control cells. After calcimycin treatment, the pS19 to total WT MYL9-FLAG ratio was significantly higher in NRMT1 KO HCT116 cells, as compared with control cells (Figure 4E,F). This confirms that Nα-acetylation is permissive for S19 phosphorylation, a modification that is critical for proper regulation of cytoskeletal dynamics, while Nα-methylation can be inhibitory.
Nα-methylation of MYL9 promotes its nuclear function
We next looked at the role of Nα-PTMs in regulating the nuclear function of MYL9, where it serves as a transcriptional activator of ICAM1 [29]. As NRMT1 is found in the nucleus, and Nα-methylation has been shown to facilitate protein–DNA interactions, we proposed that Nα-methylation of MYL9 would facilitate its transcriptional activation of ICAM1 [20,37,42–44]. To test this, we treated nontransduced as well as WT and mutant MYL9-FLAG transduced HCT116 cells with TNFα in order to induce ICAM1 transcription, and then measured the induction of ICAM1 transcription for each cell line by qPCR. We found that cells expressing the Nα-methylation enriched SPK MYL9 mutant had significantly greater induction of ICAM1 transcription than all other cell lines (Figure 5A).
Figure 5. Nα-methylation promotes the nuclear function of MYL9.
(A) Cells expressing the Nα-methylation enriched SPK mutant of MYL9 show significantly greater transcription of ICAM1 than all other TNFα stimulated cell lines. Results are shown as fold-change compared with untreated, nontransduced cells, and one-way ANOVA with Tukey’s multiple comparisons test was performed for statistical analysis (P < 0.001; Control — 9.03 ± 1.50, WT — 6.35 ± 1.36, SPK — 23.71 ± 3.21, SSQ — 4.18 ± 1.23). (B) The induction of ICAM1 transcription by SPK MYL9 was significantly decreased in methylation-deficient NRMT1 KO cells. Results are shown as fold-change compared with treated control cells, and a Student’s two-tailed t-test was performed for statistical analysis (P < 0.005; Control — 1.00, NRMT1 KO — 0.46 ± 0.11). (C) All MYL9-Dendra2 protein variants showed equal increases in nuclear localization after TNFα treatment (P < 0.05). Control Dendra2 protein showed no change in localization upon treatment. Two-way ANOVA with Bonferroni’s multiple comparisons test was used for analysis of results (Dendra2-only no treatment (No Tx) — 0.59 ± 0.01, TNFα — 0.58 ± 0.01; WT No Tx — 0.41 ± 0.005, TNFα — 0.50 ± 0.04; SPK No Tx — 0.42 ± 0.03, TNFα — 0.50 ± 0.01; SSQ No Tx — 0.41 ± 0.005, TNFα — 0.48 ± 0.005). Scale bars are 100 μM. (D) ChIP experiments showing the Nα-methylation enriched SPK MYL9-FLAG mutant was the only protein to result in detectable enrichment of the ICAM1 promoter sequence over input. The PLK promoter served as an off-target control sequence. n = 3 for all experiments. All error bars represent standard deviation.
To confirm that enrichment of Nα-methylation was responsible for the heightened transcriptional activity of SPK MYL9, and not loss of Nα-acetylation, this mutant was transduced into control and NRMT1 KO HCT116 cells and ICAM1 transcription was again assayed by qPCR. Loss of Nα-methylation reduced the transcriptional activity of SPK MYL9, as seen by a decrease in ICAM1 transcription when expressed in NRMT1 KO cells as compared with control cells (Figure 5B). This confirms a requirement for Nα-methylation to promote the optimal transcriptional activity of MYL9 in its specialized nuclear role.
We next sought to clarify the mechanism by which Nα-methylation facilitates the transcriptional activity of MYL9. First, we used immunofluorescence to determine if Nα-methylation of MYL9 results in increased nuclear localization, which would increase the pool of MYL9 available to participate in transcription. HCT116 cells transduced with WT, SPK, or SSQ MYL9-Dendra2 were imaged before and post-TNFα treatment. A Dendra2-only expressing cell line was included as a control for nonspecific changes in localization. Dendra2-only did not show TNFα responsive localization (Figure 5C). All variants of MYL9 showed a significant increase in the nuclear to total ratio of MYL9-Dendra after TNFα treatment (Figure 5C), but there were no significant differences between MYL9 variants with regards to this increase. This experiment indicates that Nα-methylated MYL9 does not increase the pool of nuclear MYL9, and that the mechanism by which this PTM facilitates transcriptional function occurs downstream of nuclear localization.
Previous studies have shown Nα-methylation to be important for the association of chromatin regulating and DNA damage response proteins with DNA [20,42–44]. To see if Nα-methylation similarly facilitates the protein–DNA interaction of MYL9 with the ICAM1 promoter, we performed ChIP in nontransduced and WT, SPK, and SSQ MYL9-FLAG expressing HCT116 cells. Only the Nα-methylation enriched SPK mutant of MYL9 resulted in enriched immunoprecipitation of the ICAM1 promoter over input (Figure 5D). This indicates that Nα-methylation facilitates the protein–DNA interaction of MYL9 with the ICAM1 promoter, and provides a mechanism by which Nα-methylation supports the transcriptional function of MYL9.
Regulation of MYL9 protein–protein interactions by Nα-acetylation and Nα-methylation
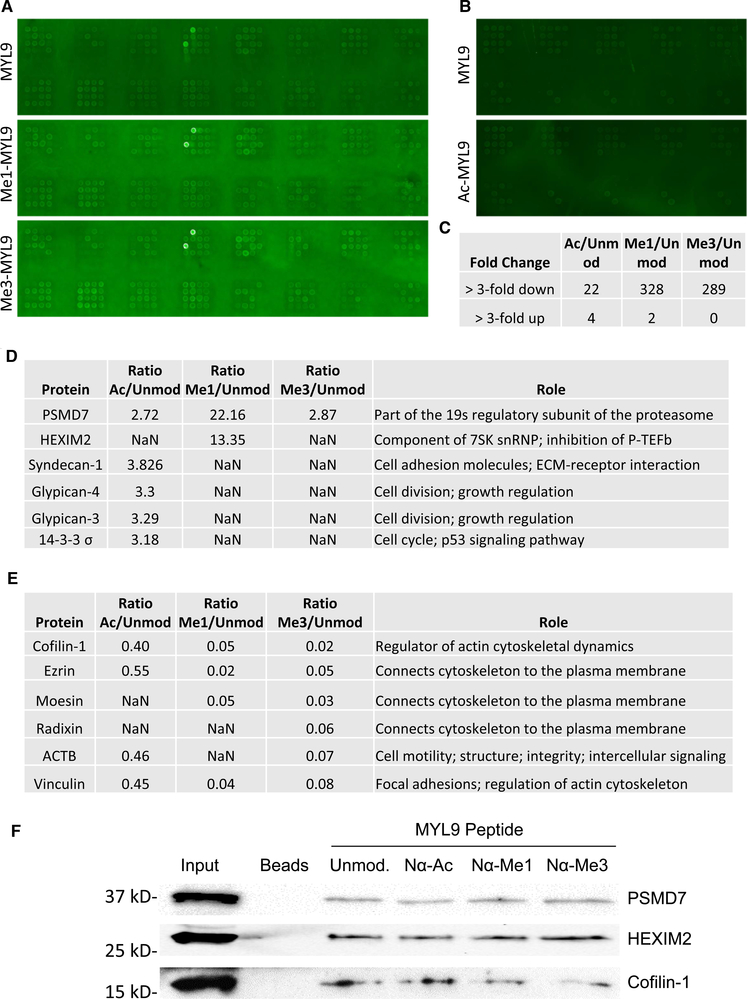
As regulating protein–protein interactions is another important general function of both acetylation and methylation, we wanted to next determine if Nα-acetylation and Nα-methylation play a role in the protein–protein interactions of MYL9. It has been recently shown that protein domains that recognize histone methylation can also recognize methylation of nonhistone proteins [9,10]. To address whether any known reader domains recognize and bind the methylated or acetylated alpha-amino group of MYL9, an in vitro binding screen was performed with arrays of immobilized recombinant methyl or acetyl reader domains [49].
The methyl reader array consisted of 104 unique domain constructs (Supplementary Figure S2A) and the acetyl reader array consisted of 30 unique domain constructs (Supplementary Figure S2B). Biotinylated N-terminal peptides of unmodified, mono, and trimethylated MYL9 were tested for binding on the methyl reader array. Unmodified and acetylated MYL9 peptides were tested for binding on the acetyl reader array. No known methyl or acetyl reader domains showed specific affinity for modified MYL9 peptides (Figure 6A,B), indicating Nα-methylated and Nα-acetylated MYL9 are read by unique domains or are not read in a domain-specific way. While this result is specific to MYL9, it seems possible that it may hold true for Nα-PTMs in general, as the N-terminal α-amine presents a considerably different steric environment than is found at the ε-amine of lysine or at the guanidino group of arginine.
Figure 6. Protein–protein interactions of Nα-acetyl- and Nα-methyl-MYL9.
Nα-PTMs of MYL9 were not specifically bound by any of the tested (A) methyl or (B) acetyl reader domains. Nonspecific binding was observed for all forms of MYL9 peptide to the Tudor domain from the LBR protein, which is consistently recognized nonspecifically. (C) The SILAC LC–MS/MS screen identified few proteins whose interactions increased with Nα-modified MYL9. However, both Nα-mono and trimethylation led to widespread blocking of interactions. (D) Only six proteins showed up-regulated interaction with Nα-modified MYL9. (E) Many of the hundreds of proteins that showed down-regulated interaction with Nα-methylated MYL9 are associated with cytoskeletal regulation. (F) Western blots of pulldowns did not confirm the increased interactions with PSMD7 and HEXIM2, but did confirm the decrease in Cofilin-1 interaction with methylated MYL9.
In attempt to identify unique protein domains that specifically recognize and bind Nα-methylation and Nα-acetylation, an unbiased stable isotope labeling of amino acids in cell culture (SILAC) liquid chromatography-mass spectrometry (LC–MS/MS) pulldown screen was performed. For pulldowns, 14 amino acid N-terminal peptides of MYL9 were synthesized in unmodified, Nα-acetylated, Nα-monomethylated, and Nα-trimethylated states. Pulldowns with unmodified MYL9 peptide were performed using protein lysate harvested from cells grown in light media. Separate pulldowns were performed with acetylated, monomethylated, or trimethylated MYL9 peptides using heavy isotope labeled protein lysate. Elutions from each modified peptide pulldown were mixed with an elution from an unmodified peptide pulldown to generate three samples that each contained light proteins that interacted with unmodified MYL9 and heavy proteins that interacted with one of the forms of modified MYL9. Proteins in these pools were identified and quantified by LC–MS/MS.
Each sample yielded ~1300 protein identifications, with 1611 total proteins identified across all three-sample pairs. Only proteins that had a SILAC ratio (peptides identified in both light and heavy states) were included for further analysis, resulting in ~650 proteins per sample pair. Of these 650, there were only six candidate interactors that showed increased binding to modified MYL9 (Figure 6C,D). Interestingly, Nα-modification of MYL9 predominantly reduced protein–protein interactions, and this was most striking for Nα-monomethylated and trimethylated MYL9. While Nα-acetylated MYL9 peptide only showed 22 downregulated protein interactions as compared with unmodified MYL9 peptide, mono and trimethylated MYL9 peptides each showed ~300 down-regulated interactions (Figure 6C). Interestingly, many of the genes that showed down-regulated binding to Nα-methyl-MYL9 were involved in cytoskeletal regulation (Figure 6E), which supports a model of Nα-methylation being able to block the cytoplasmic roles of MYL9. We selected up-regulated and down-regulated interactors for further validation by peptide pulldowns followed by Western blot. The increased binding of PSMD7 and HEXIM2 to Nα-acetyl-MYL9 did not reproduce but the decreased binding of Cofilin-1 to Nα-methyl-MYL9 was reproducible (Figure 6F). These results indicate that the small number of enhanced binding interactions seen with modified MYL9 as compared with unmodified MYL9 may have been artifactual and that blocking protein–protein interactions, as opposed to promoting protein–protein interactions, may be an important regulatory feature of MYL9 Nα-methylation.
Discussion
Regulation of stability has historically been considered the primary role for Nα-methylation and Nα-acetylation. Early studies showed that alpha-amino dimethylproline blocked degradation of cytochrome c by aminopeptidases [38,39]. This function has been proposed as a general function of Nα-methylation but has yet to been confirmed with any additional substrates [16]. More recently, a novel protein degradation pathway termed the Ac/N-end rule was described [33]. It was shown that in S. cerevisiae, proteins with an Nα-acetylated Met, Ala, Val, Ser, Thr, or Cys are targeted for proteolytic degradation by the E3 ubiquitin ligases Doa10 or Not4 [33,51]. This pathway was later demonstrated in human cells for the protein RGS2, with potential impact on blood pressure regulation [61]. WT MYL9 has an N-terminal Ser that can be Nα-acetylated, making it a potential target of the Ac/N-end rule pathway. However, we show here that blocking Nα-acetylation or Nα-methylation of MYL9 did not alter stability. This finding aligns with the model that Nα-PTMs modify protein stability in a context and substrate specific manner, with factors such as complex formation (i.e. sequestering the N-terminus) and secondary structure of the N-terminus contributing to regulatory outcomes [31,51].
As the NATs and NRMTs show distinct cellular compartmentalization, we tested whether Nα-acetylation and Nα-methylation regulate the distinct cytoplasmic and nuclear roles of MYL9 [18,31]. We first investigated how Nα-PTMs modulate the cytoplasmic role of MYL9 as a regulator of NMII activity and cytoskeletal dynamics by looking at the downstream cellular processes of cell adhesion and migration [22,24]. We found that exclusive Nα-acetylation led to both increased cell spreading on fibronectin and increased migration. These results are indicative of increased cytoskeletal dynamics occurring downstream of heightened NMII activation. We also found that exclusive Nα-acetylation of MYL9 increases the occurrence of pS19, offering a potential mechanism for the observed increase in cytoskeletal activity.
Taken together with the protein–protein interaction data, we hypothesize that Nα-acetylation is more permissive for interaction with MYL9 kinases than Nα-methylation. Precedent for the interplay of Nα-PTMs with internal residue PTMs has been set by studies of Histone H4 [62,63], and blocking of phosphorylation by methylation of a neighboring residue is commonly seen and termed a ‘methylation-phosphorylation switch’ [59,60]. Serine 19 phosphorylation of MYL9 is often accompanied by phosphorylation of threonine 18, a PTM that further increases NMII activity, and it will be interesting to see if phosphorylation of Thr 18 is also affected [27,64]. The proximity of these PTMs, taken together with our finding of interplay between Nα-modifications and pS19, suggest that the N-terminal PTMs of MYL9 may exert complex, combinatorial control over NMII activity. Future studies will further investigate the interplay between N-terminal and internal side chain PTMs of MYL9 and how Nα-modifications affect the interaction between MYL9 and its kinases and phosphatases.
While Nα-methylation appears to block protein–protein interactions, we found that similar to lysine and arginine methylation, it promotes protein–DNA interactions. Upon TNFα treatment, expression of the SPK MYL9 mutant results in significantly greater ICAM1 transcription as compared with WT or SSQ MYL9. As Nα-trimethylation results in a pH-insensitive positive charge at the amino terminus [16,18,37], and many NRMT substrates require Nα-methylation for proper recruitment to DNA and subsequent downstream functions [20,42–44], we performed ChIP and found that Nα-methylation enhances the association of MYL9 with the ICAM1 promoter. MYL9 has also been described to interact with the promoter and activate transcription of xanthine oxidase in cardiomyocytes [65], and was the only gene differentially up-regulated with age in injured rat arteries, indicating Nα-methylation of MYL9 may play an important role in cardiovascular aging as well as inflammation [66].
We have shown that Nα-methylation favors the specialized, nuclear function of MYL9 by increasing association with DNA. We have also shown that Nα-acetylation promotes the cytoplasmic functions of MYL9, at least in part, by permitting phosphorylation of Ser19. Whether Nα-acetylation actively promotes pS19, or it is actually blocked by Nα-methylation, remains to be seen. Understanding the endogenous distribution of these proteoforms will be an integral part of answering this question. If Nα-acetyl-MYL9 and Nα-methyl-MYL9 are static, distinct, and have equal access to both the cytoplasm and the nucleus, than Nα-methylation could mark a pool of MYL9 for nuclear rather than cytoplasmic activity in part by blocking protein interactions necessary for cytoplasmic function. However, if the modifications are interchangeable and dictated by cellular localization, then each could only promote its specialized compartmental function.
If the modifications are distinct, static pools, it seems levels of MYL9 Nα-acetylation would regulate substrate availability for Nα-methylation. NATs occur in the cytoplasm and catalyze both co-translational and PTM of substrates [31]. This means that NATs would have access to substrates before the NRMTs, which are compartmentalized in the nucleus, and suggests that the degree of MYL9 Nα-acetylation will dictate the degree of MYL9 left available for Nα-methylation [18,37]. In this way, Nα-acetylation could mark MYL9 molecules designated for cytoplasmic activity. In addition to MYL9, many predicted dual NRMT and NAT substrates are known to have both cytoplasmic and nuclear roles, including inversin and NEMO [67,68]. This raises the distinct possibility that regulation of compartmental activity is a conserved role for Nα-acetylation and Nα-methylation. If the modifications are interchangeable and dictated by cellular localization, cytoplasmic Nα-acetylation would have to be removed before Nα-methylation in the nucleus. This would require an alpha-amino deacetylase, and though alpha-amino deacetylases and demethylases have been proposed, both have yet to be identified [18]. However, whether or not Nα-PTMs prove to be interchangeable or static, our study makes clear that one substrate can occur as multiple Nα-proteoforms. These proteoforms have distinct functional readouts and serve to increase protein functional diversity.
The importance of uncovering the properties of protein regulation by Nα-PTMs is highlighted by developmental defects and disease states associated with dysregulation of Nα-methylation and Nα-acetylation. NRMT1 KO mice display improper development, premature aging, and premature morbidity [41]. Additionally, many mutations in NRMT1 that decrease catalytic activity have been associated with cancers, and knockdown of this enzyme has been shown to increase mammary tumorigenesis [40,45]. A mutation in NAA10, the catalytic subunit of NatA, results in the lethal X-linked developmental disorder Ogden syndrome [69,70]. Dysregulation of Nα-acetylation is also observed in multiple cancers, and overexpression of NatA has been shown to drive oncogenic transformation in lung cells [71]. Interestingly, aberrant post-translational regulation of MYL9 has also been observed in multiple cancers, including those of the breast and lung [23]. Further studies that reveal the regulatory roles of Nα-acetylation and Nα-methylation at the protein level are essential to expanding our understanding of basic cell biology and will also reveal novel pathways that contribute to disease related processes.
Supplementary Material
Acknowledgements
The authors thank Dr. Mark Bedford at the University of Texas MD Anderson Cancer Center for the peptide array work and MS Bioworks for processing and analysis of the SILAC samples.
Funding
This work was supported by a research grant from the National Institutes of Health to C.E.S.T. [GM112721] and the protein array analysis was supported by a CPRIT Grant to Dr. Mark Bedford [RP130432].
Abbreviations
- BME
basement membrane extract
- FBS
fetal bovine serum
- ICAM1
intracellular adhesion molecule 1
- KO
knockout
- MLCK
myosin light chain kinase
- MOI
multiplicity of infection
- MYL9
Myosin regulatory light chain 9
- NAA10
NatA catalytic subunit
- NAT
N-terminal acetyltransferase
- NMII
nonmuscle myosin II
- NRMT
N-terminal RCC1 methyltransferase
- PTM
post-translational modification
- RT
room temperature
- SILAC
stable isotope labeling of amino acids in cell culture
- TNFα
tumor necrosis factor alpha
- WT
wild type
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Rothbart SB and Strahl BD (2014) Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839, 627–643 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolffe AP and Hayes JJ (1999) Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720 10.1093/nar/27.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeder JE, Kwak YT, McNamara RP, Forst CV and D’Orso I (2015) HIV tat controls RNA Polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. eLife 4, e08955 10.7554/eLife.08955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boisvert FM, Rhie A, Richard S and Doherty AJ (2005) The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4, 1834–1841 10.4161/cc.4.12.2250 [DOI] [PubMed] [Google Scholar]
- 5.Boisvert FM, Dery U, Masson JY and Richard S (2005) Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19, 671–676 10.1101/gad.1279805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT et al. (2010) Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol 17, 617–619 10.1038/nsmb.1797 [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D et al. (2014) AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159, 558–571 10.1016/j.cell.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taverna SD, Li H, Ruthenburg AJ, Allis CD and Patel DJ (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol 14, 1025–1040 10.1038/nsmb1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kachirskaia I, Shi X, Yamaguchi H, Tanoue K, Wen H, Wang EW et al. (2008) Role for 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling. J. Biol. Chem 283, 34660–34666 10.1074/jbc.M806020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E et al. (2010) The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J. Biol. Chem 285, 37725–37732 10.1074/jbc.M110.139527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stützer A et al. (2014) Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53, 49–62 10.1016/j.molcel.2013.10.030 [DOI] [PubMed] [Google Scholar]
- 12.Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I et al. (2009) H3k27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 19, 221–233 10.1101/gr.080861.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ et al. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. U.S.A 107, 21931–21936 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L et al. (2010) Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 38, 4958–4969 10.1093/nar/gkq244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A et al. (2011) H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 7, e1001354 10.1371/journal.pgen.1001354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stock A, Clarke S, Clarke C and Stock J (1987) N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 220, 8–14 10.1016/0014-5793(87)80866-9 [DOI] [PubMed] [Google Scholar]
- 17.Brown JL and Roberts WK (1976) Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nα-acetylated. J. Biol. Chem 251, 1009–1014 [PubMed] [Google Scholar]
- 18.Tooley JG and Schaner Tooley CE (2014) New roles for old modifications: emerging roles of N-terminal post-translational modifications in development and disease. Protein Sci. 23, 1641–1649 10.1002/pro.2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polevoda B and Sherman F (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol 325, 595–622 10.1016/S0022-2836(02)01269-X [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF and Macara IG (2007) N-terminal α-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol 9, 596–603 10.1038/ncb1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, Balsbaugh JL, Shabanowitz J et al. (2012) Substrate specificity of mammalian N-terminal α-amino methyltransferase NRMT. Biochemistry 51, 5942–5950 10.1021/bi300278f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti MA and Adelstein RS (2008) Nonmuscle myosin II moves in new directions. J. Cell Sci. 121, 11–18 10.1242/jcs.007112 [DOI] [PubMed] [Google Scholar]
- 23.Newell-Litwa KA, Horwitz R and Lamers ML (2015) Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Dis. Model Mech 8, 1495–1515 10.1242/dmm.022103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicente-Manzanares M, Ma X, Adelstein RS and Horwitz AR (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol 10, 778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelstein RS and Conti MA (1975) Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature 256, 597–598 10.1038/256597a0 [DOI] [PubMed] [Google Scholar]
- 26.Newell-Litwa KA, Badoual M, Asmussen H, Patel H, Whitmore L and Horwitz AR (2015) ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J. Cell Biol 210, 225–242 10.1083/jcb.201504046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T et al. (1996) Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271, 20246–20249 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- 28.Yuen SL, Ogut O and Brozovich FV (2009) Nonmuscle myosin is regulated during smooth muscle contraction. Am. J. Physiol. Heart Circ. Physiol 297, H191–H199 10.1152/ajpheart.00132.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q and Sarna SK (2009) Nuclear myosin II regulates the assembly of preinitiation complex for ICAM-1 gene transcription. Gastroenterology 137, 1051–1060.e3 10.1053/j.gastro.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank PG and Lisanti MP (2008) ICAM-1: role in inflammation and in the regulation of vascular permeability. Am. J. Physiol. Heart Circ. Physiol 295, H926–H927 10.1152/ajpheart.00779.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aksnes H, Drazic A, Marie M and Arnesen T (2016) First things first: vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci. 41, 746–760 10.1016/j.tibs.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 32.Dikiy I and Eliezer D (2014) N-terminal acetylation stabilizes N-terminal helicity in lipid- and micelle-bound α-synuclein and increases its affinity for physiological membranes. J. Biol. Chem 289, 3652–3665 10.1074/jbc.M113.512459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang CS, Shemorry A and Varshavsky A (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 10.1126/science.1183147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monda JK, Scott DC, Miller DJ, Lydeard J, King D, Harper JW et al. (2013) Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure 21, 42–53 10.1016/j.str.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnesen T, Starheim KK, Van Damme P, Evjenth R, Dinh H, Betts MJ et al. (2010) The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol. Cell. Biol 30, 1898–1909 10.1128/MCB.01199-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petkowski JJ, Bonsignore LA, Tooley JG, Wilkey DW, Merchant ML, Macara IG et al. (2013) NRMT2 is an N-terminal monomethylase that primes for its homologue NRMT1. Biochem. J 456, 453–462 10.1042/BJ20131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M et al. (2010) NRMT is an α-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature 466, 1125–1128 10.1038/nature09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettigrew GW and Smith GM (1977) Novel N-terminal protein blocking group identified as dimethylproline. Nature 265, 661–662 10.1038/265661a0 [DOI] [PubMed] [Google Scholar]
- 39.Smith GM and Pettigrew GW (1980) Identification of N,N-dimethylproline as the N-terminal blocking group of Crithidia oncopelti cytochrome c557. Eur. J. Biochem 110, 123–130 10.1111/j.1432-1033.1980.tb04847.x [DOI] [PubMed] [Google Scholar]
- 40.Bonsignore LA, Butler JS, Klinge CM and Schaner Tooley CE (2015) Loss of the N-terminal methyltransferase NRMT1 increases sensitivity to DNA damage and promotes mammary oncogenesis. Oncotarget 6, 12248–12263 10.18632/oncotarget.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonsignore LA, Tooley JG, Van Hoose PM, Wang E, Cheng A, Cole MP et al. (2015) NRMT1 knockout mice exhibit phenotypes associated with impaired DNA repair and premature aging. Mech. Ageing Dev 146–148, 42–52 10.1016/j.mad.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai Q, Fu L, Wang Z, Gan N, Dai X and Wang Y (2014) α-N-methylation of damaged DNA-binding protein 2 (DDB2) and its function in nucleotide excision repair. J. Biol. Chem 289, 16046–16056 10.1074/jbc.M114.558510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sathyan KM, Fachinetti D and Foltz DR (2017) α-amino trimethylation of CENP-A by NRMT is required for full recruitment of the centromere. Nat. Commun 8, 14678 10.1038/ncomms14678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai X, Otake K, You C, Cai Q, Wang Z, Masumoto H et al. (2013) Identification of novel α-n-methylation of CENP-B that regulates its binding to the centromeric DNA. J. Proteome Res 12, 4167–4175 10.1021/pr400498y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shields KM, Tooley JG, Petkowski JJ, Wilkey DW, Garbett NC, Merchant ML et al. (2017) Select human cancer mutants of NRMT1 alter its catalytic activity and decrease N-terminal trimethylation. Protein Sci. 26, 1639–1652 10.1002/pro.3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen T, Brownawell AM and Macara IG (2004) Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol. Cell. Biol 24, 6608–6619 10.1128/MCB.24.15.6608-6619.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCaffrey LM and Macara IG (2009) The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 23, 1450–1460 10.1101/gad.1795909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H and Unser M (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176 10.1002/cyto.a.20022 [DOI] [PubMed] [Google Scholar]
- 49.Espejo A and Bedford MT (2004) Protein-domain microarrays. Methods Mol. Biol 264, 173–181 10.1385/1-59259-759-9:173 [DOI] [PubMed] [Google Scholar]
- 50.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F et al. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 10.1016/j.cell.2010.08.020 [DOI] [PubMed] [Google Scholar]
- 51.Shemorry A, Hwang CS and Varshavsky A (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 10.1016/j.molcel.2013.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF et al. (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol 24, 461–465 10.1038/nbt1191 [DOI] [PubMed] [Google Scholar]
- 53.Adam V, Nienhaus K, Bourgeois D and Nienhaus GU (2009) Structural basis of enhanced photoconversion yield in green fluorescent protein-like protein Dendra2. Biochemistry 48, 4905–4915 10.1021/bi900383a [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Gurskaya NG, Merzlyak EM, Staroverov DB, Mudrik NN, Samarkina ON et al. (2007) Method for real-time monitoring of protein degradation at the single cell level. BioTechniques. 42, 446–450 10.2144/000112453 [DOI] [PubMed] [Google Scholar]
- 55.Potter DA, Tirnauer JS, Janssen R, Croall DE, Hughes CN, Fiacco KA et al. (1998) Calpain regulates actin remodeling during cell spreading. J. Cell Biol 141, 647–662 10.1083/jcb.141.3.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huveneers S, Truong H, Fassler R, Sonnenberg A and Danen EH (2008) Binding of soluble fibronectin to integrin α5 β1 - link to focal adhesion redistribution and contractile shape. J. Cell Sci 121, 2452–2462 10.1242/jcs.033001 [DOI] [PubMed] [Google Scholar]
- 57.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM and Burridge K (1998) Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol 141, 539–551 10.1083/jcb.141.2.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nigro E, Schettino P, Polito R, Scudiero O, Monaco ML, De Palma GD et al. (2018) Adiponectin and colon cancer: evidence for inhibitory effects on viability and migration of human colorectal cell lines. Mol. Cell. Biochem 10.1007/s11010-018-3319-7 [DOI] [PubMed] [Google Scholar]
- 59.Sabbattini P, Sjoberg M, Nikic S, Frangini A, Holmqvist PH, Kunowska N et al. (2014) An H3K9/S10 methyl-phospho switch modulates Polycomb and Pol II binding at repressed genes during differentiation. Mol. Biol. Cell 25, 904–915 10.1091/mbc.e13-10-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biggar KK and Li SS (2015) Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol 16, 5–17 10.1038/nrm3915 [DOI] [PubMed] [Google Scholar]
- 61.Park SE, Kim JM, Seok OH, Cho H, Wadas B, Kim SY et al. (2015) Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 347, 1249–1252 10.1126/science.aaa3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fulton MD, Zhang J, He M, Ho MC and Zheng YG (2017) Intricate effects of α-amino and lysine modifications on arginine methylation of the N-terminal tail of histone H4. Biochemistry. 56, 3539–3548 10.1021/acs.biochem.7b00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A and Kirmizis A (2013) N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS Genet. 9, e1003805 10.1371/journal.pgen.1003805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heissler SM and Sellers JR (2014) Myosin light chains: teaching old dogs new tricks. Bioarchitecture 4, 169–188 10.1080/19490992.2015.1054092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YS, Liu B, Luo XJ, Zhang JJ, Li NS, Ma QL et al. (2015) A novel function of nuclear nonmuscle myosin regulatory light chain in promotion of xanthine oxidase transcription after myocardial ischemia/reperfusion. Free Radic. Biol. Med 83, 115–128 10.1016/j.freeradbiomed.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 66.Shehadeh LA, Webster KA, Hare JM and Vazquez-Padron RI (2011) Dynamic regulation of vascular myosin light chain (MYL9) with injury and aging. PLoS ONE 6, e25855 10.1371/journal.pone.0025855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nürnberger J, Bacallao RL and Phillips CL (2002) Inversin forms a complex with catenins and N-cadherin in polarized epithelial cells. Mol. Biol. Cell 13, 3096–3106 10.1091/mbc.e02-04-0195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang TT, Wuerzberger-Davis SM, Wu ZH and Miyamoto S (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115, 565–576 10.1016/S0092-8674(03)00895-X [DOI] [PubMed] [Google Scholar]
- 69.Myklebust LM, Van Damme P, Støve SI, Dörfel MJ, Abboud A, Kalvik TV et al. (2015) Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum. Mol. Genet 24, 1956–1976 10.1093/hmg/ddu611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Damme P, Støve SI, Glomnes N, Gevaert K and Arnesen T (2014) A Saccharomyces cerevisiae model reveals in vivo functional impairment of the Ogden syndrome N-terminal acetyltransferase NAA10 Ser37Pro mutant. Mol. Cell Proteomics 13, 2031–2041 10.1074/mcp.M113.035402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalvik TV and Arnesen T (2013) Protein N-terminal acetyltransferases in cancer. Oncogene 32, 269–276 10.1038/onc.2012.82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.