Abstract
Mammalian cells release extracellular vesicles (EVs) into their microenvironment that travel the entire body along the stream of bodily fluids. EVs contain a wide range of biomolecules. The transported cargo varies depending on the EV origin. Knowledge of the origin and chemical composition of EVs can potentially be used as a biomarker to detect, stage, and monitor diseases. In this paper, we demonstrate the potential of EVs as a prostate cancer biomarker. A Raman optical tweezer was employed to obtain Raman signatures from four types of EV samples, which were red blood cell- and platelet-derived EVs of healthy donors and the prostate cancer cell lines- (PC3 and LNCaP) derived EVs. EVs’ Raman spectra could be clearly separated/classified into distinct groups using principal component analysis (PCA) which permits the discrimination of the investigated EV subtypes. These findings may provide new methodology to detect and monitor early stage cancer.
Extracellular vesicles (EVs)1−3 are small spherical particles (diameter between 30 nm and 1 μm) enclosed by a phospholipid bilayer, shed by living cells into their extracellular environment.2 Both healthy and unhealthy cells secrete EVs so that EVs are found in all body fluids, such as blood plasma,4 urine,5 and breast milk.6 These small particles play a significant role in both intercellular communication and waste control.2,7
EVs are formed through several biogenesis pathways, for example, the endolysosomal pathway or budding from the plasma membrane.3 The vesicles formation process allows the parent cells to package biomolecules with the generated EVs, such as membrane lipids, proteins, receptors, and genetic information.3 These biomolecules are transported by the EVs from the parent cell to a recipient cell.2,4,8,9 The molecular composition of the transported cargo has been shown to change depending on the origin of the EVs. Therefore, EVs released from healthy and diseased cells are likely to contain different combinations of biological molecules. The different types of cargo in turn implies that EVs can be utilized as a disease biomarker2,3 and clinical relevance of EVs2,3 have been explored in various studies.10
Recently it was shown that EVs secreted by tumor cells contain tumor antigens.11,12 Various biochemical compositions of cancer derived EVs suggests a potential of EVs as a biomarker not only for cancer diagnosis but also for cancer prognosis and the monitoring of patients after or during treatment.13 Furthermore, if the alterations in EV molecular content can be reflected into an altered spectral behavior, then spectroscopy could be used for the analysis. Raman spectroscopy is an analytical tool long used to determine molecular composition without external labels. Therefore, vibrational spectroscopic technique presents a potentially useful opportunity for such an analysis.14−17
Spontaneous Raman spectroscopy is a type of vibrational spectroscopy technique based on inelastic scattering by molecules. When incident photons are scattered by molecules, some are scattered with particular energy shifts, a phenomenon called Raman scattering.18 Raman microscopy is used to investigate structural and compositional information on a specimen.18,19 Since the optical technique yields the fingerprint of chemicals, it has been widely used in biological and pharmaceutical fields.10,20−22 It has been applied to identify differences in tissues and cells. Convincing spectral differences have been demonstrated between cancer cells and healthy cells, based on lipid droplet content, carotenoids, and ratio between different proteins.14,21−25
Raman microscopy creates an image of the molecular composition and structure of a sample.18−20 In Raman scattering, some of the photons incident on a molecule are inelastically scattered, with the energy change of the emitted photons related to the energy states possible for the scattering molecule.18 Raman scattering does not require external labels and has been widely used in the biological and pharmaceutical fields.10,20−22,26,27 Convincing spectral differences have been demonstrated between cancer cells and healthy cells, based on lipid droplet content, carotenoids, EVs, and ratio between different proteins.14,17,21−25
Therefore, Raman spectroscopy is a promising tool to reveal the structural differences among EVs of various origins. However, the vibrational differences across the EV subtypes is subtle. Such subtle differences require sensitive and reliable analysis, such as principal component analysis (PCA). This statistical technique is used to interpret high dimensional data with several intercorrelated variables.28 PCA is widely utilized in pattern recognition, image processing, and spectroscopy. PCA differs from supervised learning in the sense that all variation is evaluated unsupervised so that dependence on peculiarities of the assignments in the training set are avoided as all spectra are used without assignment
In this study, spontaneous Raman18 was utilized to obtain spectral fingerprints of four different EVs subsets that had been derived from two prostate cancer cell lines (LNCaP and PC3) and from platelet and red blood cells from healthy donors. We obtained the spectral fingerprints of each EV subtype and used PCA to identify the four vesicles subtypes based on 300 spectra. The discrimination that we aim for is not between EVs from healthy prostate cells and EVs from cancer prostate cells since this is not a discrimination that would be useful in diagnosis (a healthy person lacks EVs from cancers cells). Rather we seek to discriminate EVs from prostate cancer cells from EVs derived from (healthy) platelets and red blood cells.
Experimental Section
Preparation of Blood Cells-Derived EVs
Red blood cell concentrate (150 mL) obtained from Sanquin (Amsterdam, The Netherlands) was diluted 1:1 with filtered phosphate-buffered saline (PBS; 154 mM NaCl, 1.24 mM Na2HPO4·2H2O, 0.2 mM NaH2PO4·2H2O, pH 7.4; supplemented with 0.32% trisodium-citrate; 0.22 mm filter (Merck Chemicals BV, Darmstadt, Germany)) and centrifuged three times for 20 min at 1 560g, 20 °C using a Rotina 46RS centrifuge (Hettich, Tuttlingen, Germany). The EV-containing supernatant was pooled, and aliquots of 50 μL were frozen in liquid nitrogen and stored at −80 °C.
Platelet concentrate (100 mL) obtained from Sanquin (Amsterdam, The Netherlands) was diluted 1:1 with filtered PBS. Next, 40 mL acid of citrate dextrose (ACD; 0.85 M trisodiumcitrate, 0.11 M d-glucose, and 0.071 M citric acid) was added and the suspension was centrifuged for 20 min at 800g, 20 °C. Thereafter, the supernatant was centrifuged (20 min at 1 560g, 20 °C). This centrifugation procedure was repeated twice to ensure removal of platelets. The vesicle-containing supernatant was pooled, and aliquots of 50 μL were frozen in liquid nitrogen and stored at −80 °C. Samples were thawed on melting ice for 30 min before use.
Preparation of Prostate Cancer-Derived EVs
Two prostate cancer cell lines (PC3 and LNCaP) were used as a model to produce prostate cancer-derived EVs. Cell lines were cultured at 37 °C and 5% CO2 in Dulbecco’s modified Eagle medium, RPMI 1640 with l-glutamine (Thermo Fischer Scientific, 11875) supplemented with 10% v/v fetal bovine serum, 10 units/mL penicillin, and 10 μg/mL streptomycin. Medium was refreshed every second day. When cells reached 80–90% confluence, they were washed three times with PBS and FBS-free RPMI medium supplemented with 1 unit/mL penicillin and 1 μg/mL streptomycin was added to the cells. After 48 h of cell culture, cell supernatant was collected and centrifuged at 1000g for 30 min. The invisible pellet containing dead or apoptotic cells and the biggest in size population of EVs was discarded. The supernatant was pooled, and aliquots of 50 μL were frozen in liquid nitrogen and stored at −80 °C. Size distribution and presence of the harvested EVs was assessed with nanoparticle tracking analysis (NTA), and transmission electron microscopy (TEM) images were taken to provide some examples of EVs.
Size Distribution Measurement Using Nanoparticle Tracking Analysis
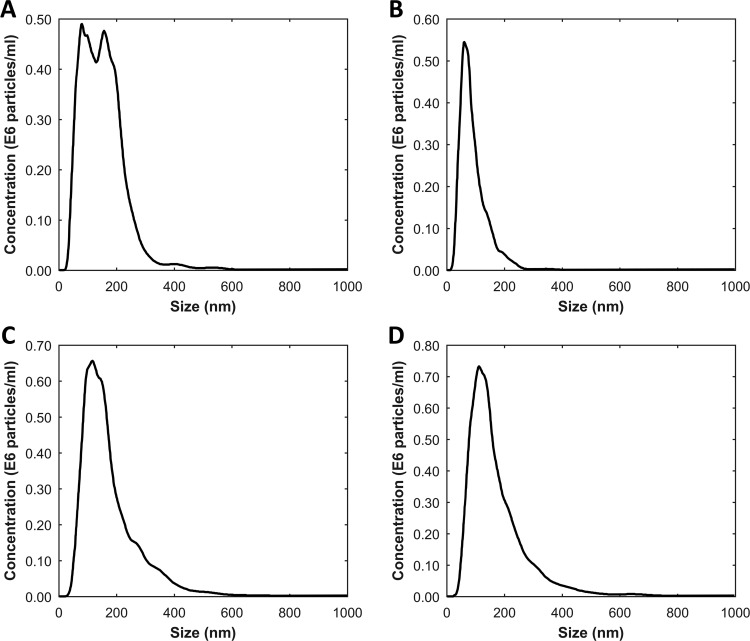
The concentration and size distribution of particles in the EV-containing samples were measured by NTA (NS500; Nanosight, Amesbury, U.K.), equipped with an EMCCD camera and a 405 nm diode laser. Silica beads (105 nm diameter; Microspheres-Nanospheres, Cold Spring, NY) were used to configure and calibrate the instrument. Fractions were diluted 10 to 2 000-fold in filtered PBS to reduce the number of particles in the field of view below 200/image. Of each sample, 10 videos, each of 30 s duration, were captured with the camera shutter set at 33.31 ms and the camera gain set at 400. All samples were analyzed using the same threshold, which was calculated by custom-made software (MATLAB v.7.9.0.529). Analysis was performed by the instrument software (NTA 2.3.0.15). Size distribution and concentration of EV samples are shown in Figure 1.
Figure 1.
Concentration and size distribution of EV samples measured using NTA. Panels A, B, C, and D represent the NTA result of red blood cell-derived EVs, platelet derived-EVs, PC3-derived EVs, and LNCaP derived-EVs, respectively. The mean size of red blood cell derived EVs is 148 ± 3.7 nm, and its concentration is 0.85 × 108 ± 0.03 × 108 particles/mL. Platelet-derived EVs is 89 ± 4.6 nm and 0.42 × 108 ± 0.02 × 108 particles/mL. PC3-derived EVs is 172 ± 3.7 nm and 1.00 × 108 ± 0.03 × 108 particles/mL. LNCaP-derived EVs is 167 ± 4.4 nm and 1.06 × 108 ± 0.05 × 108 particles/mL.
Visualizing Prepared Sample Using Transmission Electron Microscopy
Size exclusion chromatography was used to isolate EVs from the platelet and red blood cell EV-containing samples.29 Sepharose CL-2B (30 mL, GE Healthcare; Uppsala, Sweden) was washed with PBS containing 0.32% trisodiumcitrate (pH 7.4, 0.22 mm filtered). Subsequently, a frit was placed at the bottom of a 10 mL plastic syringe (Becton Dickinson (BD), San Jose, CA)), and the syringe was stacked with 10 mL of washed sepharose CL-2B to create a column with 1.6 cm in diameter and 6.2 cm in height. Platelet or red blood cell EV-containing samples (125 μL) were loaded on the respective column, followed by elution with PBS/0.32% citrate (pH 7.4, 0.22 mm filtered). The first 1 mL was discarded and the following 500 μL was collected.
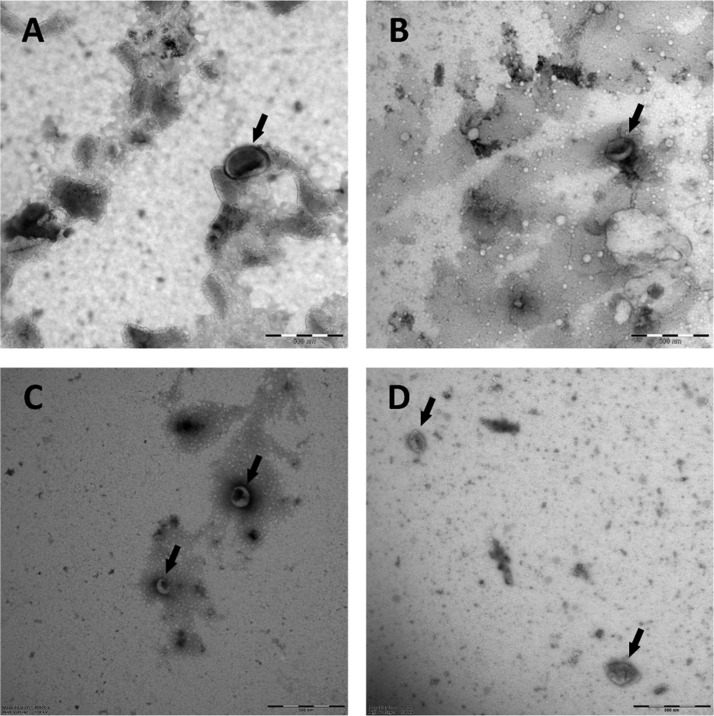
All EV samples were fixed 1:1 in a 0.1% final concentration (v/v) paraformaldehyde (Electron Microscopy Science, Hatfield, PA) for 30 min. Then, a 300 mesh carbon-coated Formvar film nickel grid (Electron Microscopy Science) was placed on 10 μL of fixed sample for 7 min. Thereafter, the grid was transferred onto drops of 1.75% uranyl acetate (w/v) for negative staining, blotted after 7 min and air-dried. Each grid was studied through a transmission electron microscope (Fei, Tecnai-12; Eindhoven, The Netherlands) operated at 100 kV using a Veleta 2,048 × 2,048 side-mounted CCD camera and Imaging Solutions software (Olympus, Shinjuku, Tokyo, Japan). All steps were performed at room temperature and all used liquids were filtered through 0.22 μm filters (Merck, Darmstadt, Germany). TEM images of the various groups of EV are shown in Figure 2.
Figure 2.
Transmission electron microscope images of EV subtypes. Arrows point EVs in the figure. (A) red blood cell-derived EVs, (B) platelet-derived EVs, (C) PC3-derived EVs, and (D) LNCaP-derived EVs. Scale bar in each panel is 500 nm.
Raman Spectral Data Acquisition
For the Raman experiments, a 50 μL volume of each EV sample was placed in a hollow cavity on a microscope slide which is made with borosilicate glass. The cavity was covered by a thin glass disk (0.25 μm, borosilicate glass to prevent evaporation and contamination.
To obtain the spectral information on EVs, a custom-built Raman microscope was used. This microscope has a Kr+ laser (Innova 90-K, Coherent Inc., Santa Clara, CA) with a wavelength of 647 nm as the excitation source. The excitation beam was focused onto the prepared sample. The scattered photons were collected by the same objective lens (40×/0.95NA UPLSAPO, Olympus Corp., Tokyo, Japan), focused on a 15 μm pinhole at the entrance of custom-made spectrograph dispersing in the range of 646–849 nm.30 The pinhole allows us to achieve confocal configuration with lateral laser spot size of about 350 nm and axial resolution of about 1.5 μm. The spectral data were dispersed by a prism based custom built spectrograph and recorded by an EMCCD camera which was cooled down to −70 °C (Newton DU-970N-BV, Andor Technology Ltd., Belfast, Northern Ireland).30
EVs are very small and float in suspension. Optical trapping allows the capturing of vesicles at the waist of the highly focused beam.10,31 We focused the excitation beam 50 μm below the bottom of the disk coverslip to minimize artifacts from the surroundings. The power of the excitation beam was 50 mW under the objective. The exposure time per spectrum was 10 and 16 spectra were obtained at the fixed position (160 s in total). After each data acquisition, we closed the shutter of the laser and moved the sample stage to allow new vesicles to be captured. Uniform experimental conditions were applied during all the experiments. We obtained 300 data sets from four different EV subtypes (75 data sets from each subtype).
Data Processing and Principal Component Analysis
All programs were implemented in MATLAB R2016b (version 9.1.0, The MathWorks, Natick, MA). Cosmic rays and the background from the microscope system were corrected. The raw Raman signal was recorded as a function of the pixel number. Pixel numbers were converted into the wavenumber scale using toluene peaks and ArHg lines (520, 785, 1003, 1030, 1210, 1604, 2919, 3056, 1097, 1303, 1705, 1910, 2126, 2145, 2357, 2508, 2873, 2964, 2977, 3114, 3131, 3354, 3560, and 3582 cm–1) for calibration.
Since the volume of EVs are about 100 folds smaller than confocal volume, the contribution of the vesicles to the total signal was much weaker than the background from the suspension (PBS or RPMI-1640 cell culture medium). To retrieve the contribution of EVs, we obtained the spectral information from pure PBS and cell culture medium and subtracted this from the collected spectral data of the EV samples. To reduce the noise, we averaged 16 spectra in a data set. Nevertheless, the processed data still contained several sources of noise, such as the offset, bending, and autofluorescence contributions. We applied baseline correction using the msbackadj with default value which is a function of the Bioinformatics Toolbox of MATLAB (see Figure 3).
Figure 3.

Raman spectra of each vesicle EV subtypes. Left column of the figure shows the untreated Raman data and curves in the right column shows preprocessed data. For the data processing, background subtraction and baseline correction were conducted. (A and E) Spectrum of red blood cell-derived EVs, (B and F) spectrum of platelet-derived EVs, (C and G) spectrum of PC3-derived EVs, and (D and H) spectrum of LNCaP-derived EVs.
For the multivariate analysis, we selected the spectral fingerprint region of each EV subtype which is the range of 400–1800 cm–1. The preprocessed spectra were normalized using unity-based normalization (feature scaling) for PCA. This brings all values into the range of 0 to 1, which prevents the emergence of artifacts as a result of variations in the intensity. PCA was performed with the function in MATLAB.
Results and Discussion
Figure 4 shows the averaged Raman spectra of EV subtypes in 400–1800 (left, fingerprint region) and 2700–3050 cm–1 (right, high-frequency region) after baseline correction and normalization. Data preprocessing was required due to the weak Raman contribution of EVs. Spectral features were observed in both the fingerprint and the high frequency region. Figure 4 shows the spectral differences across the EV subtypes (670–770, 998, 1146–1380, 1504–1590, and 1710–1780 cm–1 in the fingerprint; 2834–2897 and 1985–3025 cm–1 in the high-frequency region). Each EV subtype showed distinctive spectral features; for example, lipid contents at 2847 and 2876 cm–1, protein contribution at 2932 cm–1, CH2 deformation in lipids at 1296 cm–1, CH2 and CH3 deformation in proteins and lipids at 1440 cm–1, phenylalanine at 1603 cm–1, amide II at 1544 cm–1 and C=C stretching in lipids 1650 cm–1.32−34
Figure 4.
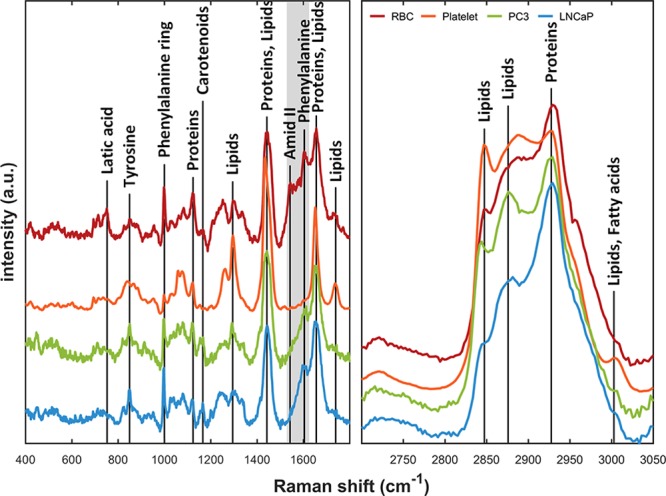
Raman spectra of EV subtypes. The curves are normalized using feature scaling method to enable comparison of the spectra in same scale. Each EV fingerprint shows spectral differences across the fingerprint area. Shaded area shows the main contribution to the separation by PCA. The spectra are vertically segregated for clarity purpose. High-frequency region (right) also shows small discrepancies between EV subtypes.
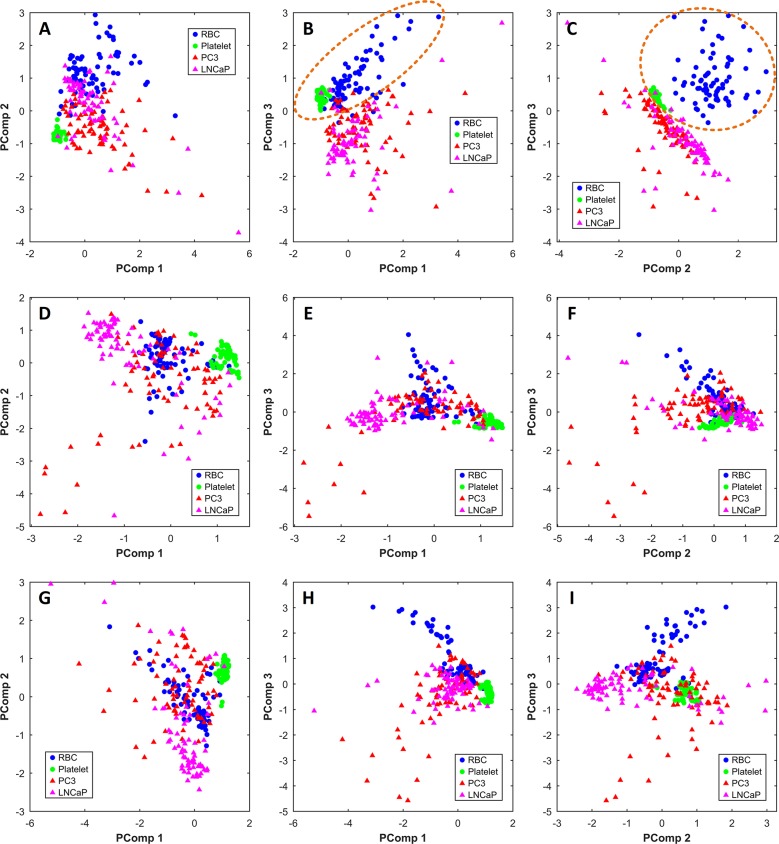
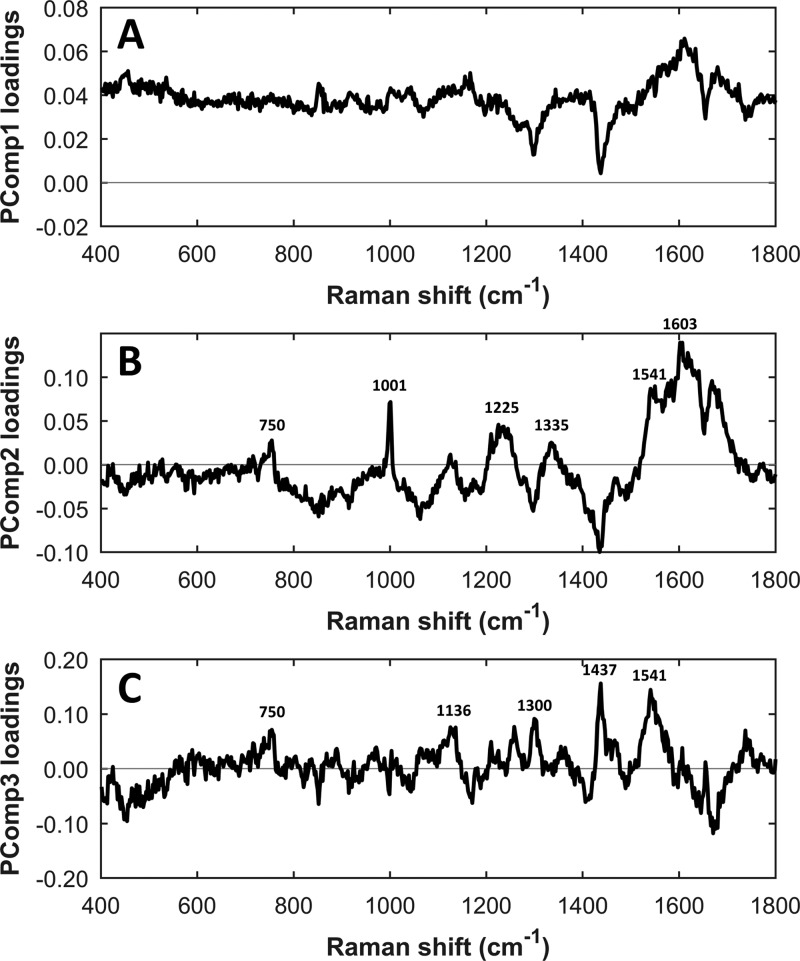
Multivariate analysis using PCA was conducted on the Raman spectra of four different EV subtypes (red blood cell-, platelet-, PC3-, and LNCaP-derived EVs). The Raman spectra in the range of 400 to 1800 cm–1 with a 2 cm–1 interval (n = 300; 654 data points) were selected for PCA. We also performed PCA in the high frequency region (2700 to 3050 cm–1) and full spectra (400 to 3050 cm–1), but these resulted in a weak separation (see Figure 5D–I). PCA in the fingerprint region performed better. The PCA score plot of PComp1 (87.47% of data variance) vs PComp2 (5.27% of data variance), PComp3 (1.36% of data variance) vs PComp1, and PComp3 vs PComp2 are shown in parts A, B, and C of Figure 5, respectively. Hematopoietic cell-derived EVs are marked with circles and cancer-derived EVs with triangles. In Figure 5B,C, PCA score plots clearly separate the prostate cancer EV group from the healthy EV group with 94.67% and 98%, respectively, of the data being accurately classified. This result indicates the clear discrimination of these two groups based on their spectral fingerprints. PCA loading plots show that those classification might be contributed by a peak at 750 cm–1 (latic acid) and a spectral band between 1500 and 1700 cm–1, which contains contribution of phenylalanine at 1603 cm–1 and amide II at 1544 cm–1 (see Figure 6).
Figure 5.
PCA score plots for the Raman spectra obtained from four EV subtypes (red blood cell-EVs, blue ●; platelet-EVs, green ●; PC3-EVs, red ▲ ; and LNCaP-EVs, pink ▲). Circles represent blood cell-derived EVs and triangles show cancer-derived EVs. Panels A–C were performed on the fingerprint region (400–1800 cm–1), panels D–F were performed on the high-frequency region (2700–3050 cm–1), and panels G–I were performed on the full spectrum (400–3050 cm–1). Panels B and C show good separation among EVs with various cellular origins. Principal component 1 (PComp1), PComp2, and PComp3 account for 87.47%, 5.27%, and 1.36% of total variance, respectively. (A) Score plot for PComp1 and Pcomp2, (B) score plot for PCom1 and PComp3, and (C) score plot for PComp2 and PComp3. In panels B and C, 94.67% and 98% of the data is classified into two categories, respectively, one containing the healthy cell-derived EVs and the other one the prostate cancer-derived EVs.
Figure 6.
PCA loading plots corresponding to (A) PComp1, (B) PComp2, and (C) PComp3.
However, some EVs were not well separated. This could be caused by the heterogeneous nature of cancer EVs and the low signal-to-noise ratio of EV spectra. Despite these limitations, the result of multivariate analysis suggests the need for further study on EVs detection and recognition for disease monitoring.
Conclusion
In conclusion, we have explored spectral differences between cancer-derived EVs and healthy control-derived EVs to examine the potential of cancer-derived EVs as a cancer biomarker. To clarify the role of EVs as a disease biomarker, Raman spectroscopy was employed to obtain the spectral fingerprint of EV subtypes. We obtained 300 data sets from four EV subtypes, and Raman spectra of EVs were analyzed with PCA to classify vesicle subtypes. Based on principal component of the spectral information, the result of multivariate analysis shows the spectral differences between healthy cells derived EVs (red blood cell and platelet) and prostate cancer cell-derived EVs (PC3 and LNCaP). The PC score plot shows that more than 90% of EVs were classified into two categories. This result suggests the potential of EVs as a cancer biomarker and makes them worthy for further investigation.
Acknowledgments
This work is part of the research program (Cancer-ID) with Project Number 14197, which is financed by The Netherlands Organization for Scientific Research (NWO).
Author Contributions
⊗ A.N. and L.R. contributed equally.
The authors declare no competing financial interest.
References
- Park J. O.; Choi D. Y.; Choi D. S.; Kim H. J.; Kang J. W.; Jung J. H.; Lee J. H.; Kim J.; Freeman M. R.; Lee K. Y. Proteomics 2013, 13, 2125–2134. 10.1002/pmic.201200323. [DOI] [PubMed] [Google Scholar]
- van der Pol E.; Böing A. N.; Harrison P.; Sturk A.; Nieuwland R. Pharmacol. Rev. 2012, 64, 676–705. 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- Verma M.; Lam T. K.; Hebert E.; Divi R. L. BMC Clin. Pathol. 2015, 15, 6. 10.1186/s12907-015-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caby M.-P.; Lankar D.; Vincendeau-Scherrer C.; Raposo G.; Bonnerot C. Int. Immunol. 2005, 17, 879–887. 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- Pisitkun T.; Shen R.-F.; Knepper M. A. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 13368–13373. 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Admyre C.; Johansson S. M.; Qazi K. R.; Filén J.-J.; Lahesmaa R.; Norman M.; Neve E. P.; Scheynius A.; Gabrielsson S. J. Immunol. 2007, 179, 1969–1978. 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- Denzer K.; Kleijmeer M. J.; Heijnen H.; Stoorvogel W.; Geuze H. J. J. Cell Sci. 2000, 113, 3365–3374. [DOI] [PubMed] [Google Scholar]
- Beach A.; Zhang H.-G.; Ratajczak M. Z.; Kakar S. S. J. Ovarian Res. 2014, 7, 14. 10.1186/1757-2215-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C.; Melo S. A.; Protopopov A.; Tang J.; Seth S.; Koch M.; Zhang J.; Weitz J.; Chin L.; Futreal A. J. Biol. Chem. 2014, 289, 3869–3875. 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatischeff I.; Larquet E.; Falcón-Pérez J. M.; Turpin P.-Y.; Kruglik S. G. J. Extracell. Vesicles 2012, 1, 19179. 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre F.; Schartz N. E.; Movassagh M.; Flament C.; Pautier P.; Morice P.; Pomel C.; Lhomme C.; Escudier B.; Le Chevalier T. Lancet 2002, 360, 295–305. 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- Rabinowits G.; Gerçel-Taylor C.; Day J. M.; Taylor D. D.; Kloecker G. H. Clin. Lung Cancer 2009, 10, 42–46. 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- Coumans F.; Doggen C. J. M.; Attard G.; De Bono J.; Terstappen L. W. M. M. Annals of oncology 2010, 21, 1851–1857. 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- Song X.; Airan R. D.; Arifin D. R.; Bar-Shir A.; Kadayakkara D. K.; Liu G.; Gilad A. A.; van Zijl P. C.; McMahon M. T.; Bulte J. W. Nat. Commun. 2015, 6, 6719. 10.1038/ncomms7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudiger C. W.; Min W.; Saar B. G.; Lu S.; Holtom G. R.; He C.; Tsai J. C.; Kang J. X.; Xie X. S. Science 2008, 322, 1857–1861. 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljković M.; Chernenko T.; Romeo M. J.; Bird B.; Matthäus C.; Diem M. Analyst 2010, 135, 2002–2013. 10.1039/c0an00042f. [DOI] [PubMed] [Google Scholar]
- Carney R. P.; Hazari S.; Colquhoun M.; Tran D.; Hwang B.; Mulligan M. S.; Bryers J. D.; Girda E.; Leiserowitz G. S.; Smith Z. J.; Lam K. S. Anal. Chem. 2017, 89, 5357–5363. 10.1021/acs.analchem.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman C. V. Indian Journal of Physics 1928, 2, 387–398. [Google Scholar]
- Kneipp K.; Kneipp H.; Itzkan I.; Dasari R. R.; Feld M. S. Chem. Rev. 1999, 99, 2957–2976. 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- Vankeirsbilck T.; Vercauteren A.; Baeyens W.; Van der Weken G.; Verpoort F.; Vergote G.; Remon J. P. TrAC, Trends Anal. Chem. 2002, 21, 869–877. 10.1016/S0165-9936(02)01208-6. [DOI] [Google Scholar]
- Austin L. A.; Osseiran S.; Evans C. L. Analyst 2016, 141, 476–503. 10.1039/C5AN01786F. [DOI] [PubMed] [Google Scholar]
- Kast R. E.; Tucker S. C.; Killian K.; Trexler M.; Honn K. V.; Auner G. W. Cancer Metastasis Rev. 2014, 33, 673–693. 10.1007/s10555-013-9489-6. [DOI] [PubMed] [Google Scholar]
- Haka A. S.; Shafer-Peltier K. E.; Fitzmaurice M.; Crowe J.; Dasari R. R.; Feld M. S. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 12371–12376. 10.1073/pnas.0501390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong Y. H.; Lim M.; Liu Q. Opt. Express 2012, 20, 22158–22171. 10.1364/OE.20.022158. [DOI] [PubMed] [Google Scholar]
- Huang Z.; McWilliams A.; Lui H.; McLean D. I.; Lam S.; Zeng H. Int. J. Cancer 2003, 107, 1047–1052. 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- Park J.; Hwang M.; Choi B.; Jeong H.; Jung J.-H.; Kim H. K.; Hong S.; Park J.-H.; Choi Y. Anal. Chem. 2017, 89, 6695–6701. 10.1021/acs.analchem.7b00911. [DOI] [PubMed] [Google Scholar]
- Bryce D. A.; Kitt J. P.; Harris J. M. J. Am. Chem. Soc. 2018, 140, 4071–4078. 10.1021/jacs.7b13777. [DOI] [PubMed] [Google Scholar]
- Abdi H.; Williams L. J. Wiley interdisciplinary reviews: computational statistics 2010, 2, 433–459. 10.1002/wics.101. [DOI] [Google Scholar]
- Böing A. N.; van der Pol E.; Grootemaat A. E.; Coumans F. A. W.; Sturk A.; Nieuwland R. J. Extracell. Vesicles 2014, 3, 23430. 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker L.Microspectroscopic Characterisation of Gold Nanorods for Cancer Cell Detection. Ph.D. Thesis, University of Twente, Enschede, the Netherlands, 2011. [Google Scholar]
- Ajito K.; Torimitsu K. Appl. Spectrosc. 2002, 56, 541–544. 10.1366/0003702021955015. [DOI] [Google Scholar]
- Smith Z. J.; Lee C.; Rojalin T.; Carney R. P.; Hazari S.; Knudson A.; Lam K.; Saari H.; Ibañez E. L.; Viitala T. J. Extracell. Vesicles 2015, 4, 28533. 10.3402/jev.v4.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramczyk H.; Surmacki J.; Kopeć M.; Olejnik A.; Lubecka-Pietruszewska K.; Fabianowska-Majewska K. Analyst 2015, 140, 2224–2235. 10.1039/C4AN01875C. [DOI] [PubMed] [Google Scholar]
- Movasaghi Z.; Rehman S.; Rehman I. U. Appl. Spectrosc. Rev. 2007, 42, 493–541. 10.1080/05704920701551530. [DOI] [Google Scholar]