Abstract
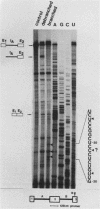
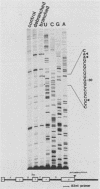
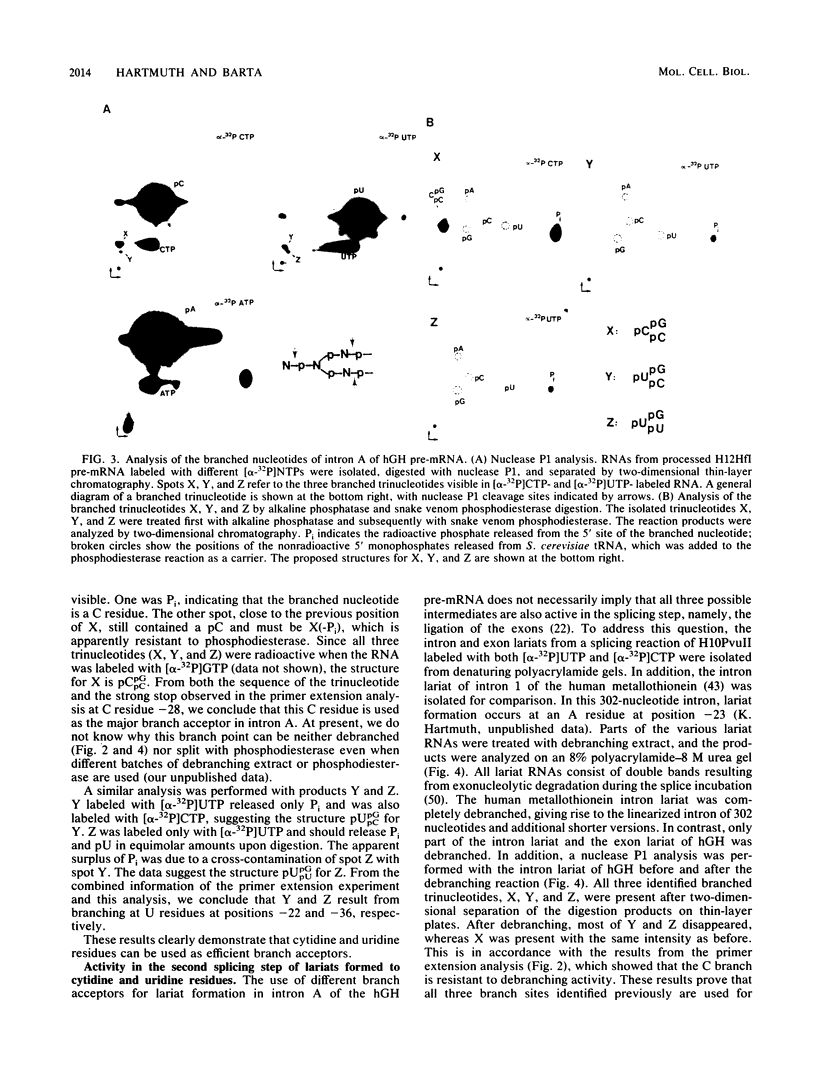
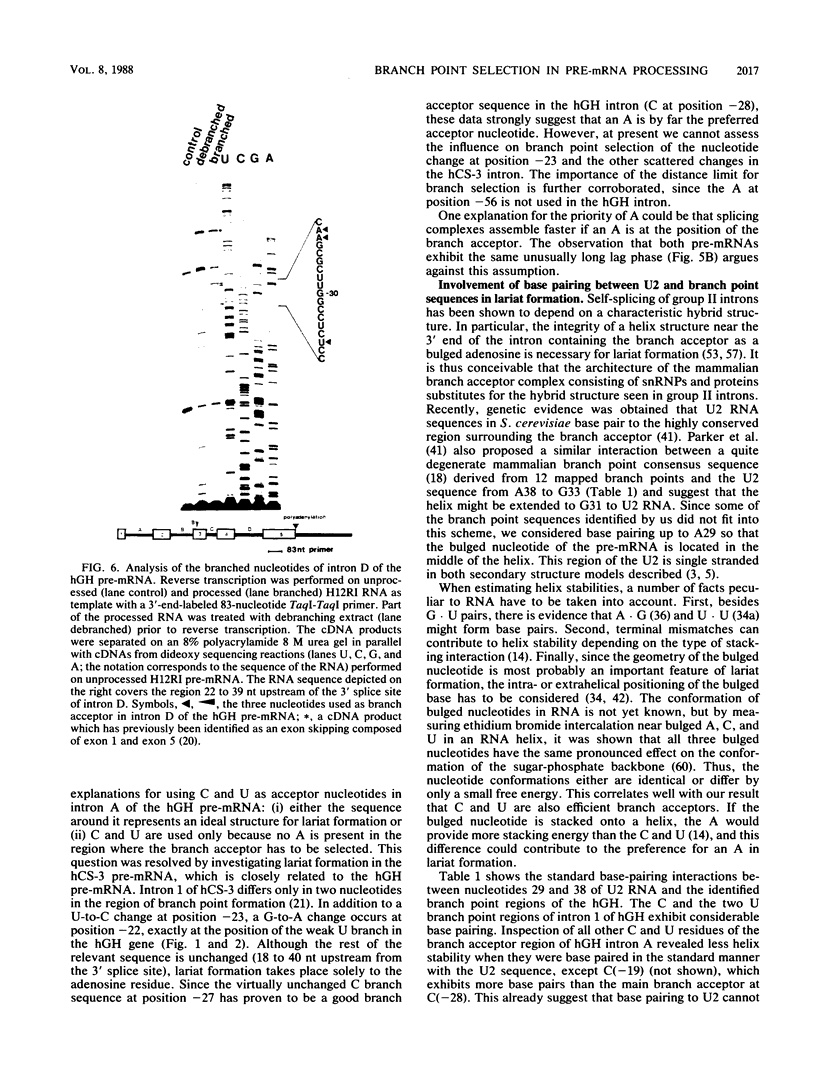
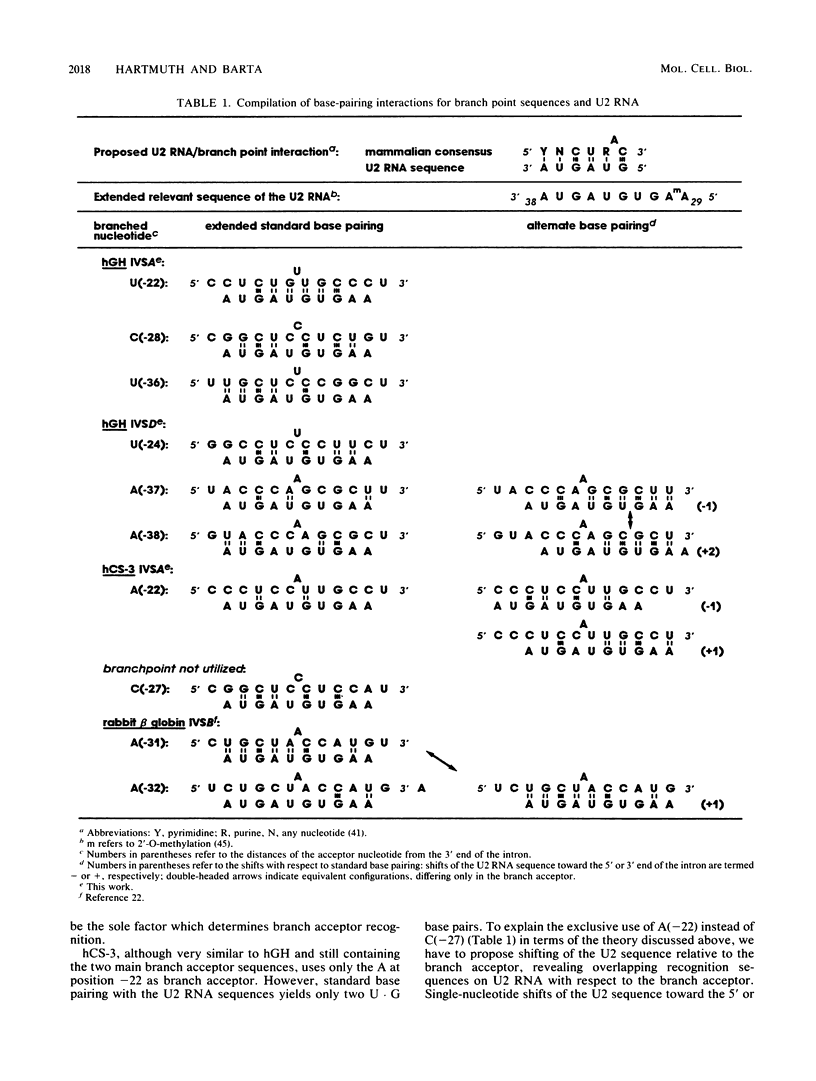
Intron A of the human growth hormone gene does not contain an A residue within 56 nucleotides preceding the 3' splice site. The analysis of the excised intron lariat revealed a C residue 28 nucleotides upstream from the 3' splice site as the major branch acceptor nucleotide. Two additional minor branched nucleotides were identified as U residues at positions -22 and -36. An adenosine substitution at position -22 results in lariat formation solely to this nucleotide. Therefore, C and U residues can function efficiently as natural branch acceptors, but an A residue is preferred if available in the proper region. In addition, the data strongly reinforce the importance of the distance constraint for lariat formation. To explain selection of the branch acceptor nucleotide, potential base-pairing interactions of branch point sequences with the U2 RNA are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Ares M., Jr U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986 Oct 10;47(1):49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Robberson B. L. U1, U2, and U4/U6 small nuclear ribonucleoproteins are required for in vitro splicing but not polyadenylation. Cell. 1986 Aug 29;46(5):691–696. doi: 10.1016/0092-8674(86)90344-2. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Black D. L., Steitz J. A. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell. 1986 Aug 29;46(5):697–704. doi: 10.1016/0092-8674(86)90345-4. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R., Cheroutre H., Degrave W., Fiers W. Simple, efficient in vitro synthesis of capped RNA useful for direct expression of cloned eukaryotic genes. Nucleic Acids Res. 1982 Oct 25;10(20):6353–6362. doi: 10.1093/nar/10.20.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Fouser L. A., Friesen J. D. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986 Apr 11;45(1):81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., Arenas J., Perkins K. K., Furneaux H. M., Pick L., Young B., Roberts R. J., Hurwitz J. In vitro formation of a lariat structure containing a G2'-5'G linkage. J Biol Chem. 1987 Mar 25;262(9):4267–4273. [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. In vitro processing of a plant pre-mRNA in a HeLa cell nuclear extract. Nucleic Acids Res. 1986 Oct 10;14(19):7513–7528. doi: 10.1093/nar/14.19.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. In vitro processing of the human growth hormone primary transcript. Nucleic Acids Res. 1987 Sep 11;15(17):7005–7025. doi: 10.1093/nar/15.17.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H., Kimelman J., Birnbaum M. J., Chen E. Y., Seeburg P. H., Eberhardt N. L., Barta A. The human growth hormone gene locus: structure, evolution, and allelic variations. DNA. 1987 Feb;6(1):59–70. doi: 10.1089/dna.1987.6.59. [DOI] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987 Jul 3;50(1):17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltwasser G., Spitzer S. G., Goldenberg C. J. Assembly in an in vitro splicing reaction of a mouse insulin messenger RNA precursor into a 60-40S ribonucleoprotein complex. Nucleic Acids Res. 1986 May 12;14(9):3687–3701. doi: 10.1093/nar/14.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel M., Eyermann F., Westhof E., Romby P., Expert-Bezançon A., Ebel J. P., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J Mol Biol. 1987 Nov 5;198(1):91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Itoh M., Okada N., Miyata T. Novel models for RNA splicing that involve a small nuclear RNA. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4471–4474. doi: 10.1073/pnas.78.7.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Aebi M., Hornig H., Weissmann C., Sharp P. A. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Patzelt E., Thalmann E., Hartmuth K., Blaas D., Kuechler E. Assembly of pre-mRNA splicing complex is cap dependent. Nucleic Acids Res. 1987 Feb 25;15(4):1387–1399. doi: 10.1093/nar/15.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. A self-splicing RNA excises an intron lariat. Cell. 1986 Jan 31;44(2):213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Riedel N., Wise J. A., Swerdlow H., Mak A., Guthrie C. Small nuclear RNAs from Saccharomyces cerevisiae: unexpected diversity in abundance, size, and molecular complexity. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8097–8101. doi: 10.1073/pnas.83.21.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Schmelzer C., Schmidt C., May K., Schweyen R. J. Determination of functional domains in intron bI1 of yeast mitochondrial RNA by studies of mitochondrial mutations and a nuclear suppressor. EMBO J. 1983;2(11):2047–2052. doi: 10.1002/j.1460-2075.1983.tb01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer C., Schweyen R. J. Self-splicing of group II introns in vitro: mapping of the branch point and mutational inhibition of lariat formation. Cell. 1986 Aug 15;46(4):557–565. doi: 10.1016/0092-8674(86)90881-0. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H. The human growth hormone gene family: nucleotide sequences show recent divergence and predict a new polypeptide hormone. DNA. 1982;1(3):239–249. doi: 10.1089/dna.1.1982.1.239. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U., Parker R., Tamm J., Iimura Y., Rossi J., Abelson J., Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986 Jul;5(7):1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. C., Edmonds M. Polyadenylylated nuclear RNA contains branches. Proc Natl Acad Sci U S A. 1983 Feb;80(4):950–954. doi: 10.1073/pnas.80.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. A., Draper D. E. Single base bulges in small RNA hairpins enhance ethidium binding and promote an allosteric transition. Nucleic Acids Res. 1987 May 26;15(10):4049–4064. doi: 10.1093/nar/15.10.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986 Jan 31;44(2):225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]